Abstract
Objective
Maternal unbalanced nutritional habits during embryonic development and perinatal stages perturb hypothalamic neuronal programming of the offspring, thus increasing obesity-associated diabetes risk. However, the underlying molecular mechanisms remain largely unknown. In this study we sought to determine the translatomic signatures associated with pro-opiomelanocortin (POMC) neuron malprogramming in maternal obesogenic conditions.
Methods
We used the RiboTag mouse model to specifically profile the translatome of POMC neurons during neonatal (P0) and perinatal (P21) life and its neuroanatomical, functional, and physiological consequences.
Results
Maternal high-fat diet (HFD) exposure did not interfere with offspring's hypothalamic POMC neuron specification, but significantly impaired their spatial distribution and axonal extension to target areas. Importantly, we established POMC neuron-specific translatome signatures accounting for aberrant neuronal development and axonal growth. These anatomical and molecular alterations caused metabolic dysfunction in early life and adulthood.
Conclusions
Our study provides fundamental insights on the molecular mechanisms underlying POMC neuron malprogramming in obesogenic contexts.
Keywords: POMC neuron, RiboTag, Neuronal programming, Obesity, Translatome
Graphical abstract
Highlights
-
•
Cell-specific POMC neuron translatome profiling in neonatal and perinatal developmental stages.
-
•
Maternal obesogenic environment alters progeny's POMC neuron translatome signatures.
-
•
Maternal HFD impairs POMC spatial distribution and axonal outgrowth, which is related to offspring's metabolic dysfunction.
-
•
The HFD intervention stage (early life or adulthood) differentially impact POMC neuron transcriptional signatures.
1. Introduction
Obesity has dramatically increased over the last 30 years and is considered one of the major health challenges of the century [1]. Epidemiological and experimental evidence show that nutritional perturbations during crucial embryonic and perinatal periods result in developmental adaptations that predispose to metabolic disturbances into adulthood [2]. In this context, it has been reported that maternal over nutrition during gestation and lactation interfere with the ontogeny of hypothalamic circuits implicated in energy balance, thus contributing to the development of obesity and type 2 diabetes (T2D) in adult offspring [3]. Pro-opiomelanocortin (POMC) neurons, located in the arcuate nucleus (ARC) of the hypothalamus, are the fundamental nexus of the melanocortin system in the regulation of appetite, energy expenditure, and metabolism [4]. The impact of gestational and/or perinatal obesogenic environments on POMC neuron programming has been illustrated by increased expression of POMC neuropeptide, impaired formation of projections to target areas, as well as altered glial-neuron communication [[5], [6], [7]]. However, the molecular mechanisms underlying POMC neuron malprogramming in maternal obesogenic conditions remain largely unknown. To exclusively target POMC neurons we implemented an in vivo Cre-mediated translating ribosome affinity purification strategy known as RiboTag [8], which allows cell-type-specific translatome profiling, avoiding artifacts derived from tissue fixation or digestion. Here we show that, in mice, maternal high-fat diet (HFD) exposure did not interfere with offspring hypothalamic POMC neuronal number, but significantly impaired their spatial distribution and axonal extension to target areas. Importantly, we established POMC neuron-specific translatome signatures accounting for such aberrant neuronal maturation. These anatomical and molecular alterations underlie the metabolic dysfunctions observed in early life and adulthood. Our study provides fundamental insights on the molecular mechanisms underlying POMC neuron malprogramming in obesogenic contexts.
2. Material and methods
2.1. RiboTag mouse line
The RiboTag mouse model allows Cre-mediated hemagglutinin (HA) epitope tagging of polyribosomes and subsequent isolation of actively translated transcriptome in a cell-specific manner [8]. The RiboTag model was crossed with POMC-Cre mice [9], generating PomcRiboTag animals. This mouse line allows translatome analysis exclusively in POMC neurons. Food and water were provided ad libitum throughout the study. Mice were maintained on a light/dark (12/12 h) and temperature-controlled conditions. All in vivo studies were performed with the approval of the University of Barcelona Ethics Committee, complying with current Spanish and European legislation.
2.2. Mouse breeding, diets, and neonatal hypothalami collection
Six-week-old PomcRiboTag female mice received either a standard chow diet (13% Kcal derived from fat; Envigo Laboratories #2014) or experimental HFD (45% Kcal derived from fat; Research Diets #D12451) during 4 weeks. After this period, the females were crossed with chow-fed PomcRiboTag male mice. Litter size was adjusted (between P1–P4) to six to eight pups to ensure adequate and standardized nutrition until weaning. The mediobasal hypothalamus of neonatal (P0) and perinatal (P21) groups were dissected and directly frozen in liquid nitrogen. Samples were kept at −80 °C until preparation for analysis.
2.3. POMC-specific ribosome-associated mRNA isolation
Isolation of Pomc mRNA was prepared as previously described with minor modifications [8]. Briefly, mediobasal hypothalami from PomcRiboTag chow and HFD offspring were ice-cold homogenized in 300 μL homogenization buffer (50 mM Tris, 100 mM KCl, 12 mM MgCl2, 1% Nonidet P-40, 1 mM DTT, 200 U/mL Promega RNasin, 1 mg/mL heparin, 100 mg/mL cycloheximide, Sigma protease inhibitor mixture at pH 7.5). After clearing, 40 μL of the homogenate was separated as INPUT sample, 350 μL of lysis buffer (RLT buffer + beta mercaptoethanol) was added, and the mixture was stored at −80 °C. The remaining homogenate was mixed with 2 μL of a mouse monoclonal anti-HA antibody (HA.11 clone 16B12, Biolegend) and placed on a gentle spinner in a cold room for 2 h. Afterward, the antibody-tissue homogenates were mixed with 200 μL of Dynabeads protein G magnetic beads (#10004G, Life Technologies) and incubated on a spinner in a cold room for another 2 h. Immunoprecipitates (IPs) were washed 3 times for 10 min with 800 μL of high-salt buffer (50 mM Tris, 300 mM KCl, 12 mM MgCl2, 1% Nonidet P-40, 1 mM DTT, 100 mg/mL cycloheximide at pH 7.5) at 4 °C in a cold room on a rotator. Then, 350 μL of lysis buffer was added to each sample immediately after removing the final high-salt wash buffer. The samples were vortexed for 30 s to break apart the antibody-bead-protein bond and placed in a magnetic stand for separation from beads. Total RNA was purified using a RNeasy Plus Micro Kit (#74034, Qiagen). Final RNA was diluted in 20 μL of RNase-free water and quantified using a Quant-iT™ RiboGreen™ RNA assay kit (#R11490, Thermo Fisher). RNA integrity (RIN) was assessed on a 2100 Bioanalyzer device (Agilent Technologies) using the RNA 6000 Pico kit (Agilent Technologies). Only samples with RIN greater than 8 were used for RNASeq analysis.
2.4. RNASeq and analysis
cDNA was synthetized from 3 to 20 ng RNA with the SMARTer Ultra Low Input RNA kit v4 (Clontech). Sequencing libraries were prepared with the NexteraXT kit (Illumina) and were sequenced in a HiSeq 2500 (Illumina) obtaining 50 base single read fragments. In total, two lanes, containing 9 and 10 samples (nine- and ten-fold multiplexing), were used and lanes were mixed with samples from all the groups. The samples passed the quality controls established in the FastQC software http://www.bioinformatics.babraham.ac.uk/projects/fastqc, were mapped to mouse reference genome mm10 with TopHat v2.0.13 [10], and its alignment was checked with Picard Tools v1.80 (available online at: http://broadinstitute.github.io/picard). Gene expression counts were obtained with the HTSeq software v.0.6.1 [11]; sample distribution was checked by Correspondence Analysis (made4 library) [12] from Bioconductor [13]; and differential gene expression was performed with the edgeR library [14]. Differentially gene expressed genes were those with a moderated p-value lower than 0.05 by False Discovery Rate, a fold change between the compared groups higher than 1.5 or lower than −1.5, and a Counts per million (CPM) value higher than 20 in at least 50% of the samples in at least one group. The differentially expressed genes were graphically represented with heat diagrams (dChip software) [15], volcano plots, and Venn diagrams (ggplot2 library [16]; https://www.bioconductor.org/; https://cran.r-project.org/web/packages/pheatmap/index.html). Pathway analysis was performed using Metacore (Clarivate).
2.5. Data visualization into networks
The differentially expressed gene (DEG) sets for the chow-fed and HFD offspring were analyzed using the BINGO tool [17] and plugged into the visualization software Cytoscape (version 3.7.1) [18], with a cutoff p-value < 0.05. Transcript network interactomes were built in Cytoscape (version 3.7.1) [18], based on the interactions previously found linking the gene ontology (GO) categories defined in this study related to neuronal migration and final anatomical positioning, and the list of transcription factors differentially expressed (Supplementary Table 1).
2.6. Genome-Wide Association Study (GWAS) extraction
A normalized list of genes corresponding to differentially expressed genes (DEGs) exclusive for the chow (659) and HFD (1984) mice were mapped to the human gene identifiers by extracting the overlapping genes from the most recent version of the “Complete List of Human and Mouse Homologs with phenotype annotations” (in www.informatics.jax.org/homology.shtml). The most recent version of the GWAS Catalog (EMBL-EBI) was used to obtain the Single Nucleotide Polymorphisms (SNPs) corresponding to the list of human identifiers associated with obesity and diabetes (gwas_catalog_v1.0.2-associations_e96_r2019-11-21.tsv; https://www.ebi.ac.uk/gwas/docs/file-downloads).
2.7. Quantitative-PCR (qPCR) analysis
Five ng of INPUT (total RNA) and immunoprecipitation (POMC neuron-specific translatome) RNA products were reverse transcribed with a SuperScript IV first strand synthesis system according to manufacturing instructions (#18090050, Thermo Fisher). Quantitative polymerase chain reactions (qPCRs) were conducted using Premix Ex Taq (#RR39WR, Takara) in the ABI Prism 7900 HT system (Applied Biosystems). Taqman Gene Expression assay FAM/TAMRA probes (Applied Biosystems) used for qPCR analysis are: Pomc (Mm00435874_m1); Agrp (Mm00475829_g1); Aldh1l1 (Mm03048957_m1); and Th (Mm00447557_m1). The level of expression was normalized against housekeeping gene GAPDH (Mm99999915_g1).
2.8. POMC neuronal counting
A detailed ontogenic analysis of POMC neurons during development until complete maturation of the anorexigenic neural circuit was performed in PomcRiboTag offspring of mothers fed with either chow or HFD. Neonatal (P0) and weaning day (P21) brains (n = 6–7 per group) were dissected and fixed in 4% parafolmaldehyde (PFA) overnight at 4 °C. Brains were further cryoprotected in 30% sucrose, frozen in smashed dry ice, and cut using a cryostat (Leica CM 1950) in four series. Brain sections (20-μm thick) of one series were blocked with 2% chicken serum in Phosphate-buffered Saline (KPBS) + 0.4% Triton X-100 and incubated with rabbit anti-POMC precursor (1:1000; #H-029-30, Phoenix Pharmaceuticals) in blocking solution overnight at 4 °C. As secondary antibody, a chicken anti-rabbit Alexa Fluor 488 (1:300; #A21441, Life Technologies) in KPBS + 0.4% Triton X-100 was used (2 h at room temperature). Images were obtained using an Olympus fluorescence microscope equipped with a 20x objective. The total number of POMC neurons per series was counted using FIJI (ImageJ) Software.
2.9. POMC and HA colocalization immunofluorescence
Cryoprotected PomcRiboTag P21 brains (n = 3) were frozen in smashed dry ice and sectioned using a cryostat (Leica CM 1950). Selected 20-μm sections (1 out of 4 sections) were blocked with 2% donkey serum in KPBS + 0.4% Triton X-100 and incubated with mouse anti-HA antibody (1:1000; HA.11 clone 16B12, Biolegend) in blocking solution overnight at 4 °C. As a secondary antibody, a donkey anti-mouse Alexa Fluor 594 (1:300; #A21202, Life Technologies) in KPBS + 0.4% Triton X-100 was used (2 h at room temperature). POMC immunostaining was performed on top of HA staining as previously described. For quantification, representative images throughout the ARC (bregma between −1.22 and −2.30 mm) of each animal were acquired using an Olympus fluorescence microscope equipped with a 20x objective. The total number of exclusive POMC+ neurons, exclusive HA+ cells, and double POMC+/HA+ was counted using FIJI (ImageJ) Software.
2.10. α-Melanocyte-stimulating hormne (MSH) immunofluorescence and fiber density quantitative analysis
Cryoprotected P21 brains (n = 3–4 per group) were frozen in smashed dry ice and sectioned using a cryostat (Leica CM 1950). Selected 20-μm sections (1 out of 4 sections) were blocked with 2% donkey serum in KPBS + 0.4% Triton X-100 and incubated with sheep anti-α-MSH (1:1000; #AB5087, Millipore) in blocking solution overnight at 4 °C. As a secondary antibody, a donkey anti-sheep Alexa Fluor 488 (1:300; #A11015, Life Technologies) in KPBS + 0.4% Triton X-100 was used (2 h at room temperature). For quantification, representative sections through the paraventricular nucleus of the hypothalamus (PVH) (bregma between −0.59 and −1.23 mm) and ARC (bregma between −1.22 and −2.30 mm) of each animal were acquired using an Olympus fluorescence microscope equipped with a 20x objective. Five images throughout the PVH and ARC of each animal were taken. A α-MSH fiber density analysis was performed using FIJI (ImageJ) Launcher based on previously published reports [19,20]. Briefly, each single image was binarized to compensate for differences in fluorescence intensity, specified in a random 200 × 200 μm region, and skeletonized, so that each fiber segment was 1 pixel thick. The integrated intensity was then measured for each image. The total density value was obtained by the sum of all image planes analyzed.
2.11. POMC neuronal spatial distribution
Tri-dimensional (3D) and statistical analysis were performed as previously described using Free-D and p-maps software [21,22]. Cryoprotected P21 brains were frozen in smashed dry ice and sectioned using a cryostat (Leica CM 1950). Selected 20-μm sections (1 out of 4 sections) were immunostained against POMC as described earlier. For POMC neuronal mapping, representative sections (one every 80 μm) throughout the ARC (bregma between −1.22 and −2.30 mm) of each animal were acquired using an Olympus fluorescence microscope equipped with a 10x objective. ARC was defined as the region with the highest neuron densities that contains 90% of the POMC population of chow-O and HFD-O groups. 3D models of POMC neuronal population were then built and spatially normalized in Free-D software for each sample. Then, POMC neuron density maps were computed for each group (n = 4–5 per group) using the same parameter for cell density estimation (rank k = 6). These maps were statistically compared, and p-value maps and iso-p-value surfaces were generated using thresholds equal to 0.05 and 0.95 (alpha = 10%) to show regions with significant cell density differences [21,22].
2.12. Physiological measurements
Body weights and gonadal fat pads were measured using a precision scale. For the glucose tolerance test, mice (n = 6–8 per group) were intraperitoneally (IP) injected with a single bolus of d-glucose (2 g/kg) after 6 h (P21 animals) or overnight (adult mice) fasting. Blood glucose levels were measured using a glucometer (Nova Pro Biomedical) after 0, 15, 30, 60, and 120 min of glucose administration. Plasma insulin levels were measured with commercially available enzyme-linked immunosorbent assay (ELISA) kits (Crystal Chem #90080).
2.13. Statistical analysis
All values are expressed as mean ± standard error of the mean (SEM). Statistical analyses were conducted using GraphPad PRISM (version 7.0). Statistical significance was determined using an unpaired two-tailed Student t test or two-way analysis of variance (ANOVA) with a Sidak multiple comparisons test as appropriate. P values less than 0.05 were considered to be statistically significant.
2.14. Data availability
The RNASeq datasets generated and/or analyzed during the current study are available in the NCBI GEO repository under accession number GSE135639. [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135639].
3. Results and discussion
3.1. Maternal HFD exposure alters offspring physiology and predisposes male offspring to obesity
To investigate the repercussion of a maternal obesogenic environment in progeny's POMC neuron development and function, we generated POMCRiboTag mice by crossing the RiboTag model [8] with a POMC-specific Cre-expressing line [9]. Six-week-old POMCRiboTag females were divided into two dietary groups: standard chow or HFD. After 4 weeks, mice were mated and maintained with the same diet until the weaning of the offspring (Figure 1A). Despite equivalent body weight throughout the study (Supplementary Fig. 1a), HFD-fed females exhibited evident metabolic dysfunction at the onset of pregnancy, such as glucose intolerance and hyperinsulinemia (Supplementary Figs. 1b–d). Metabolic and dietary changes were further illustrated by increased adiposity at the time of weaning (Supplementary Fig. 1e). These metabolic alterations confirm the implementation of a maternal obesogenic environment.
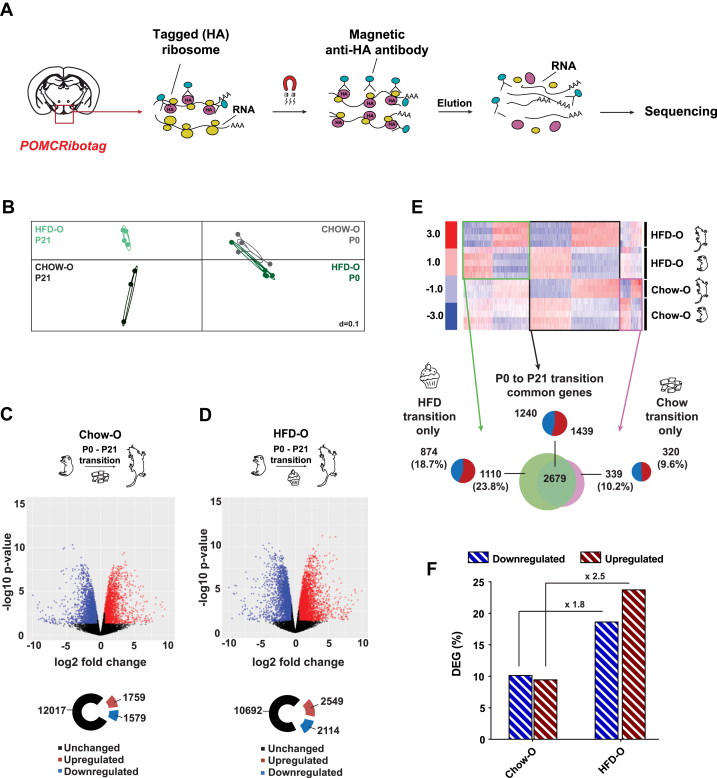
Figure 1.
Maternal HFD affects offspring physiology and predisposes male offspring to obesity. (A) Experimental design of maternal dietary conditions and offspring collection. (B–C) Body weight differences between P0 (B) and P21 (C) offspring. (D–E) Glucose tolerance test and area under the curve (AUC) of P21 male offspring (n = 6 biologically independent samples). (F) Blood glucose levels of P21 male offspring after 6 h of fasting (n = 6 biologically independent samples). (G) Body weight of 12-week-old male Chow-O and HFD-O fed with a chow diet from weaning onward (n = 16 biologically independent samples). (H) Blood glucose levels, under fed and fasting conditions, of 12-week-old male offspring (n = 6 biologically independent samples). (I–J) Glucose tolerance test and area under the curve (AUC) of 12-week-old male Chow-O and HFD-O fed with a chow diet from weaning onward (n = 10 biologically independent samples). (K) Body weight after 8 weeks of HFD exposure in Chow and HFD male offspring (n = 10 biologically independent samples). (L) Blood glucose levels, under fed or fasting conditions, after 8 weeks of HFD in Chow and HFD male offspring (n = 6–9 biologically independent samples). (M–N) Glucose tolerance test and area under the curve (AUC) after 8 weeks of HFD in Chow and HFD male offspring (n = 7–9 biologically independent samples). Data are expressed as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 ∗∗∗∗p < 0.0001.
The development of the melanocortin system in rodents can be subdivided into two major stages: (a) embryonic, when neurogenesis and specification of neuronal populations occur; and (b) postnatal, in which neurons complete maturation by establishing projections to specific target regions creating functional circuits [23]. Thus, we selected two key developmental stages to compare the effects of maternal obesogenic diet on specification [post-natal day (P)0] and maturation (P21) of POMC neurons and its effects on metabolism. HFD offspring (HFD-O) were born smaller than chow offspring (Chow-O) (Figure 1B) but presented rapid catch-up growth during lactation, becoming significantly heavier at weaning (Figure 1C; Supplementary Fig. 2a). At this age, HFD-O mice were glucose intolerant (Figure 1D–E; Supplementary Figs. 2b–c) and hyperglycemic (Figure 1F; Supplementary Fig. 2d). These abnormalities were more severe in males than females, suggesting a higher susceptibility of the former to the deleterious developmental effects of a HFD.
An obesogenic environment during crucial developmental periods exerts adverse, long-term outcomes on adult metabolic health. To confirm long-lasting metabolic effects on maternal HFD exposure, both Chow-O and HFD-O mice were weaned into a standard chow diet until adulthood. Body weight differences observed at P21 were normalized at 12 weeks of age in both male and female offspring (Figure 1G; Supplementary Fig. 2e). However, hyperglycemia and glucose intolerance persisted only in male HFD-O mice (Figure 1H–J; Supplementary Figs. 2f–h). These results confirm that maternal dietary influences on glucose homeostasis endure into male progeny's adulthood.
To examine if exposure to an obesogenic environment during embryonic and perinatal life increases obesity risk into adulthood, we challenged Chow-O and HFD-O mice with a HFD for 8 consecutive weeks. Remarkably, HFD administration during adulthood exacerbated the obese phenotype and glucose intolerance of male, but not female, HFD-O (Figure 1K-N; Supplementary Fig. 2i-l), thus confirming the predisposition to obesity acquired during intrauterine and early life.
3.2. Maternal HFD exposure alters the POMC neuron-specific translatome in the offspring
To investigate the molecular basis of POMC neuron malprogramming on maternal HFD feeding, we took advantage of the POMCRiboTag model to target actively translated mRNAs, specifically in POMC neurons, via polysome immunoprecipitation (Figure 2A) [8]. Recombination in POMC neurons was confirmed by immunofluorescence via colocalization with the HA tag (Supplementary Figs. 3a and b). Low off-target recombination was observed in the ARC that could be due to the differentiation of POMC-progenitors into AgRP neurons [24] and Kisspeptin neurons [25], or to the stochastic nature of Cre recombinase expression [26]. The reliability of the model was manifested by a marked enrichment of the Pomc gene but not of negative markers such as tyrosine hydroxylase (Th) or astrocytic aldehyde dehydrogenase 1 (Aldh1I1) (Supplementary Fig. 3c). A slight enrichment in the Agrp transcript was also observed (Supplementary Fig. 3c), which likely reflects the shared ontogenic origin of the POMC and AgRP neurons [24] and progenitor characteristic expression profile [27]. Furthermore, modest Agrp transcript expression in POMC neurons has been also observed in adult mice [28,29], suggesting that Agrp might play a role in specific subsets of the POMC neuronal population.
Figure 2.
POMC neuronal translatome signatures. (A) Schematic overview of PomcRiboTag immunoprecipitation strategy for active translatome attainment. (B) COA plot of experimental samples (n = 3–5 biologically independent samples). (C–D) Volcano plot of POMC neuronal transcript expression during P0 to P21 transition in Chow (C) and HFD (D) offspring. Threshold for fold-change (±1.5) and false discovery rate (FDR) (<0.05) was considered. DEGs during POMC neuronal maturation are depicted in blue (downregulated) and red (upregulated). Unchanged genes are represented in black (n = 3–5 biologically independent samples). (E) Heatmap illustrating DEGs during P0 to P21 transition. Diagrams show DEGs exclusive from Chow transition (in pink), exclusive from HFD transition (in green), and independent from the type of the maternal diet (in black) (n = 3–5 biologically independent samples). (F).Percentage of DEGs during POMC neuronal maturation in Chow and HFD offspring. Data are expressed as mean ± SEM.
Correspondence analysis (COA) of individual sequencing samples identified four distinct clusters representing different populations (Figure 2B). Sequencing of ribosome-associated mRNA showed differential expression of 3338 genes during P0–P21 transition in Chow-O (Figure 2C), whereas there were 4663 DEGs in HFD-O (Figure 2D). Interestingly, 2679 of the DEGs were common between both groups, showing a signature of diet-independent genes necessary for POMC neuronal maturation (Figure 2E). Moreover, out of the total amount of DEGs observed in Chow-O during P0–P21 transition, 10.2% of these genes (339) were downregulated and 9.6% (320) were upregulated (Figure 2E). On maternal HFD consumption, 18.7% of the genes were downregulated (874) and 23.8% were upregulated (1110) (Figure 2E), showing a respective 1.8- and 2.5-fold increase in exclusive-HFD DEGs in the course of POMC neuronal maturation (Figure 2F).
3.3. Maternal HFD exposure alters POMC neuron-specific developmental translatomic signatures affecting neuronal distribution and axonal extension
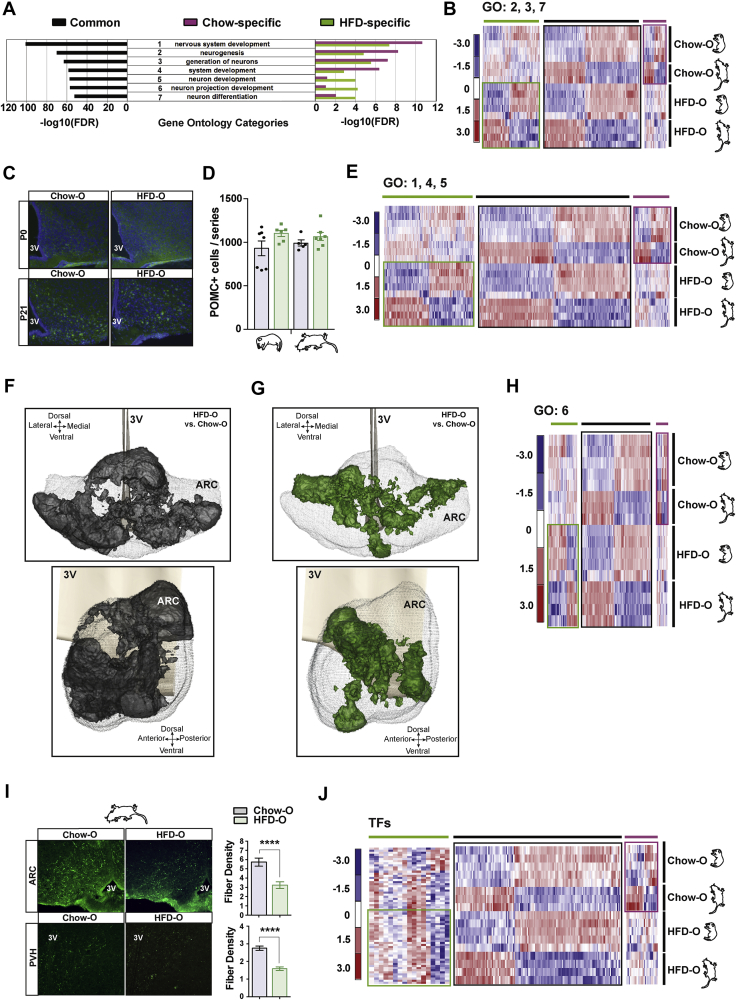
GO analysis of the significantly DEGs uncovered notable changes in key categories related to neurogenesis and neuronal differentiation, nervous system development, and neuronal projection development during intrauterine and early life (Figure 3A). To define the impact of such molecular alterations on actual POMC neuron developmental irregularities, we aimed at analyzing three main biological processes: (a) achievement of neuronal identity (comprising GO categories 2. Neurogenesis, 3. Generation of neurons, and 7. Neuron differentiation); (b) migration of neural progenitors to specific nuclei (comprising GO categories 1. Nervous system development, 4. System development, and 5. Neuron development); and (c) extension of axonal fibers to target regions (comprising GO category 6. Neuron projection development) (Figure 3A).
Figure 3.
Translatome and anatomical analysis of POMC neuronal development. (A) Gene ontology (GO) analysis of the seven top-ranked significantly altered processes during P0 to P21 POMC neuronal transition. Diagrams represent false discovery rate (FDR) for GO terms enrichment in food-independent transition (in black), Chow-exclusive transition (in pink), and HFD-exclusive transition (in green). (B) Heatmap illustrating the expression of genes within selected neuronal generation GO categories in P0 and P21 Chow-O and HFD-O. Food-independent genes (in black), Chow-exclusive genes (in pink), and HFD-exclusive genes (in green) are highlighted (n = 3–5 biologically independent samples). (C) Representative 20x images of POMC neurons in P0 and P21 Chow-O and HFD-O. (D) POMC neuronal number quantification of P0 and P21 Chow-O and HFD-O (n = 6–7 biologically independent samples). (E) Heatmap illustrating the expression of genes within selected neuronal maturation GO categories in P0 and P21 Chow-O and HFD-O. Food-independent genes (in black), Chow-exclusive genes (in pink), and HFD-exclusive genes (in green) are highlighted (n = 3–5 biologically independent samples). (F) Coronal and sagittal view of iso-p-value surface representing P21 ARC regions with significantly decreased POMC neuronal density in HFD-O compared with Chow-O (n = 4 biologically independent samples). (G) Coronal and sagittal view of iso-p-value surface representing P21 ARC regions with significantly increased POMC neuronal density in HFD-O in comparison to Chow-O (n = 4 biologically independent samples). (H) Heatmap illustrating the expression of genes within selected neuron projection development GO categories in P0 and P21 Chow-O and HFD-O. Food-independent genes (in black), Chow-exclusive genes (in pink), and HFD-exclusive genes (in green) are highlighted (n = 3–5 biologically independent samples). (I) Representative immunofluorescence 20x images showing α-MSH fiber density in the ARC and PVN of P21 Chow-O and HFD-O mice and integrated density quantification (n = 3 biologically independent samples). (J) Heatmap illustrating the expression of differentially expressed TFs in P0 and P21 Chow-O and HFD-O. Food-independent genes (in black), Chow-exclusive genes (in pink), and HFD-exclusive genes (in green) are highlighted (n = 3–5 biologically independent samples). 3 V: third ventricle. ARC: arcuate nucleus. PVH: paraventricular nucleus. Data are expressed as mean ± SEM. ∗∗∗∗p < 0.0001.
Of the 1834 genes encompassing GO categories related to neuronal differentiation, 821 (40.8%) were differentially expressed in Chow-O throughout POMC neuronal maturation, whereas 1057 (57.6%) of the genes were altered on HFD exposure (Figure 3B; Supplementary Fig. 4a). Moreover, the number of DEGs exclusive of HFD-O during P0–P21 transition demonstrated a 2.8-fold change compared Chow-exclusive DEGs. Despite the remarkable changes in the number of DEGs related to neuronal differentiation between Chow-O and HFD-O, the main functional themes and interactions within this GO category were almost identical (Supplementary Fig. 4c). Furthermore, genes indispensable for POMC identity, such as Nkx2.1 [30], Isl1 [31], Nhlh2 [32], Rbpj [33], and Tbx3 [34] did not show expression differences on HFD exposure (Supplementary Fig. 4d). In line with this, the overall number of POMC neurons was equivalent at both developmental stages (Figure 3C–D). Collectively, these results indicate that despite maternal nutrient excess, the organism can compensate and specify POMC neurons correctly.
Adequate neuronal function relies on precise anatomical location. We found 1336 (45.2%) and 1707 (57.7%) genes (Supplementary Fig. 5a) belonging to GO categories corresponding to neuronal migration and final anatomical positioning, differentially expressed in the Chow-O and HFD-O groups during P0–P21 transition, respectively. Approximately 38% of the DEGs (1120) were common between groups, whereas 7.3% (216) of the genes were exclusive of Chow-O (3.2% up- and 4.1% downregulated) and 19.8% (587) of the genes were unique for HFD-O (10.0% up- and 9.9% downregulated) (Figure 3E; Supplementary Fig. 5a). Interestingly, the fold-change increase in the number of DEGs exclusive of HFD-O compared with Chow-O exclusive ones during P0–P21 transition was similar to the neuronal identity category, with a 2.72-fold change. Next, we verified whether these translatome perturbations were actually associated with defective POMC neuron spatial organization. To this aim, we conducted a computer-based POMC neuron 3D reconstruction and spatial comparison between Chow-O and HFD-O to assess regions where POMC neurons are predominantly located [35]. The area covered by the POMC population of both groups was defined as the region of interest (ROI), delimiting the ARC. We observed a statistically significant deficit of POMC neurons in HFD-O compared with Chow-O that corresponded to 29.8% of the ROI (Figure 3F), particularly in ventrolateral portions, where POMC neurons are typically distributed [4]. Conversely, HFD-O neurons were aberrantly distributed over a smaller dorsomedial posterior ARC area corresponding to 9.6% of the ROI (Figure 3G). This was consistent with a modest decrease in POMC neuron abundance in the posterior ARC regions of HFD-O (Supplementary Fig. 4b), without significant impact in the overall POMC neuron population (Figure 3C,D). These alterations in spatial organization of POMC neurons as a consequence of maternal HFD, together with the marked decrease in POMC neuron axonal projections, must co-occur with significant cellular rearrangements. It is likely that this reorganization not only affects POMC neurons, but also other ARC neuronal subtypes and glial cells, thus altering neuronal function and connectome. These perturbations might contribute to the predisposition to develop metabolic syndrome in the offspring. Together, our results indicate that exposure to an obesogenic environment throughout intrauterine and perinatal development perturb the expression of a large subset of genes orchestrating POMC neuron migration, impairing their anatomical location albeit normal POMC neuronal differentiation.
To achieve complete maturation and function, POMC axonal afferents must reach their definitive targets. Neuron projection development was one of the main GO processes that was altered on differential dietary exposure during P0–P21 transition (Figure 3A). Translatome analysis showed 350 and 432 DEGs in Chow-O and HFD-O groups, respectively, during POMC neuron maturation. Among these, 40.4% (308 genes) were commonly altered in both groups; 5.5% of the genes (42) were exclusively changed in Chow-O; and 16.3% of the genes (124) were differentially expressed only in HFD-O (Figure 3H; Supplementary Fig. 5b), showing a 2.95-fold increase in DEGs exclusive of HFD-O during P0–P21 transition. To evaluate whether these translatome deviations led to inadequate POMC neuron axonal growth, we measured fiber density of the POMC-derived neuropeptide α-MSH in two major hypothalamic target areas: the PVH and the ARC. Consistent with previous reports [[5], [6], [7]], we observed a striking decrease in α-MSH fiber immunofluorescence in both areas (Figure 3I). These results are in agreement with our translatome data and confirm a defective regulation of POMC axonal growth to target areas due to developmental obesogenic exposure.
Transcription factors (TFs) are fundamental for neuronal programming, as they orchestrate indispensable nervous system developmental processes, including hypothalamic neural progenitor proliferation, regional patterning, cell fate determination, migration, and axonal outgrowth [36]. Metacore analysis showed 227 differentially expressed TFs, 56.4% of which (128) were altered independently of dietary conditions, whereas 12.8% (29) and 30.8% (70) were exclusive of Chow-O and HFD-O groups during P0 to P21 transition, respectively (Figure 3J). Interestingly, known players of cell fate induction and maintenance, migratory paths and axonal outgrowth, were differentially expressed during POMC maturation under obesogenic conditions (Supplementary Table 1). Thus, perturbed TF expression could underlie the defects in POMC neuron positioning and targeting characterized here.
3.4. The impact of HFD on POMC neuron-specific transcriptomic profile differs between neuronal programming and adult obesity
Next, we aimed to identify potential candidate genes whose expression may be dysregulated by fat-rich diets and thus implicated in general energy imbalance processes. To do so, we compared DEGs exclusively found during POMC neuron maturation in HFD-O with genes that changed in POMC neurons from adult mice as a result of HFD feeding [37]. Only 36 genes were common in both experimental conditions, with two thirds of the genes exhibiting expression changes in opposite directions (Figure 4A). Despite the notable differences in design between the study by Campbell et al. [37] and our study, these observations emphasize the divergent transcriptional impact of nutrient perturbations during the embryonic/perinatal period and adulthood in POMC neurons. Among the genes with a consistent direction in expression, only Nucks1 has been experimentally implicated in the hypothalamic control of energy balance. In mice, a hypothalamic deficiency of NUCKS1, a protein that modulates chromatin structure as well as replication and transcription, causes overweight and altered glucose homeostasis, likely by impairing hypothalamic insulin signaling [38].
Figure 4.
Integration of POMC translatome alterations. (A) Heatmap illustrating common DEGs found to be altered in our translatome analysis of POMC neurons during P0 to P21 transition in HFD-O and the DropSeq transcriptomic analysis of POMC clusters (Pomc/Ttr, Pomc/Anxa2, Pomc/Glipr1) in adult mice exposed to 60% HFD [37]. (B) Overlapping gene identifiers of translatome analysis of POMC neurons during P0 to P21 transition in HFD-O mapped onto obesity and type 2 diabetes GWAS-associated genes. (C) Transcript network interactome analysis showing the genomic interface between differential GO processes (neuronal migration and axonal outgrowth) and TFs during offspring's POMC neuron maturation in a maternal obesogenic context.
Finally, we analyzed whether the DEGs exclusively observed in the HFD-O group are enriched in GWAS-associated loci linked to metabolic disorders. Our analysis showed an incidental contribution of GWAS candidate genes, as only 64 (3.2%) and 33 (1.7%) of DEGs have been linked to T2D and obesity risk, respectively (Figure 4B). These results evidence the dissimilarities between the genetic mechanisms underlying obesogenic programming and common obesity. Nevertheless, the expression of these susceptibility genes in HFD-O POMC neurons were overall altered when compared with Chow-O (Figure 4B).
Our results support the notion that moderate changes in a large collection of genes, rather than dramatic perturbations of specific ones, are responsible for POMC neuronal malprogramming. For example, individual analysis of classical determinants of neuronal migration, neurite formation, and wiring of functional forebrain neurons (e.g., members of integrins, plexins, netrins, laminins, and semaphorins) [36] show modest although significant changes in gene expression. In line with this idea, network interactome analysis illustrates the overlap of an ample subset of DEGs within the GO categories for neuronal migration and axonal outgrowth, as well as for transcription factors, that might underlie POMC neuronal dysfunction on HFD exposure (Figure 4C). Hence, a collective and synergistic effect, despite modest individual expression changes, may contribute to the overall disturbances in POMC neuron anatomy and function due to obesogenic developmental exposure.
4. Conclusions
In summary, here we provide evidence that maternal obesogenic conditions, which are related to neural development, specification, and maturation processes, significantly alter distinct POMC neuron translatome signatures of the progeny. Importantly, these alterations in molecular programs are consistent with particular POMC neuron phenotypical outcomes, such as perturbed spatial distribution and impaired axonal extension to target areas, as well as systemic metabolic disturbances. Collectively, these results unveil the translatomic basis for POMC neuron malprogramming in response to maternal nutrient excess, further delineating the causes of unbalanced neuronal control of metabolism and persistent obesity risk into adulthood. Our study also raises awareness of the detrimental health effects of the current western lifestyle in offspring.
Author contributions
RH-T conceived the study, participated in its design, performed experiments, analyzed results and contributed to the manuscript preparation. JA analyzed sequencing data, contributed to the manuscript preparation and secured funding. AO performed enrichment expression analysis, neuron spatial distribution analysis using Free-D software and discussed results. JB performed neuron spatial distribution analysis using Free-D software. EES analyzed sequencing data, performed BINGO and GWAS analyses. IC, MM-G, SR, MP performed part of metabolic experiments. AGG-V set up mRNA immunoprecipitations. LAV conceived the study and participated in its design. MC conceived the study, participated in its design, coordinated and supervised the experiments, contributed to the manuscript preparation and secured funding.
Acknowledgments
We thank Ainhoa Garcia (IDIBAPS, Barcelona) for support with animal work. This study was funded by: The Bo & Kerstin Hjelt Diabetes Foundation (JA); European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement 725004) and CERCA Programme/Generalitat de Catalunya (MC). RH-T was a recipient of a FAPESP postdoctoral fellowship (2016/01868-2). AO is supported by the Beatriu de Pinós programme (2016BP00136) from the Generalitat de Catalunya and a Miguel Servet contract (CP19/00083) from Instituto de Salud Carlos III and co-financed by European Regional Development Fund (ERDF) “A way to build Europe”. SR is a recipient of Juan de la Cierva fellowship (FJCI-2016-28911) from the Spanish Ministry of Science and Innovation. MC was a recipient of a Miguel Servet 2 contract (MSII15/00025) from Instituto de Salud Carlos III and co-financed by European Regional Development Fund (ERDF) “A way to build Europe”. This work was carried out in part at Esther Koplowitz Centre.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.02.006.
Contributor Information
Roberta Haddad-Tóvolli, Email: haddad@clinic.cat.
Jordi Altirriba, Email: jorge.altirriba@unige.ch.
Arnaud Obri, Email: obri@clinic.cat.
Elena Eyre Sánchez, Email: eyre@clinic.cat.
Iñigo Chivite, Email: chivite@clinic.cat.
Maria Milà-Guasch, Email: mila@clinic.cat.
Sara Ramírez, Email: ramirez@clinic.cat.
Alicia G. Gómez-Valadés, Email: agarciag@clinic.cat.
Macarena Pozo, Email: pozo@clinic.cat.
Jasmine Burguet, Email: jasmine.burguet@inra.fr.
Licio A. Velloso, Email: lavelloso.unicamp@gmail.com.
Marc Claret, Email: mclaret@clinic.cat.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supplementary Figure 1HFD effects on maternal metabolism. (a) Maternal body weight before exposure to HFD (6 weeks of age), on the day of mating, and by the time of offspring's weaning (n = 16–24 biologically independent samples). (b-c) Maternal glucose tolerance test and area under the curve (AUC) at the onset of pregnancy (n = 8 biologically independent samples). (d) Maternal plasma insulin levels under fed and fasting conditions at the onset of pregnancy (n = 8 biologically independent samples). (e) Maternal gonadal white adipose tissue weight, relative to total body weight, in Chow and HFD females after weaning of their offspring (n = 21–29 biologically independent samples). Data are expressed as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001. Supplementary Figure 2Maternal HFD effects on female offspring metabolism. (a) Body weight at P21 in Chow and HFD female offspring (n = 6 biologically independent samples). (b-c) Glucose tolerance test and area under the curve (AUC) of P21 female offspring (n = 6 biologically independent samples). (d) Blood glucose levels at P21 of female HFD offspring after 6 h of food deprivation (n = 6 biologically independent samples). (e) Body weight in 12-week-old Chow-O and HFD-O females fed with a chow diet from weaning onward (n = 6–12 biologically independent samples). (f) Blood glucose levels, under fed and fasting conditions, of 12-week-old female HFD offspring (n = 6 biologically independent samples). (g-h) Glucose tolerance test and area under the curve (AUC) of 12-week-old Chow-O and HFD-O female offspring fed with a chow diet from weaning onward (n = 6 biologically independent samples). (i) Body weight of female Chow-O and HFD-O mice after 8 weeks of HFD exposure (n = 6 biologically independent samples). (j) Blood glucose levels under fed and fasting conditions after 8 weeks of HFD exposure in female Chow-O and HFD-O mice (n = 6 biologically independent samples). (k-l) Glucose tolerance test and area under the curve (AUC) after 8 weeks of HFD exposure in female Chow-O and HFD-O mice (n = 6 biologically independent samples). Data are expressed as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001. Supplementary Figure 3Validation of the RiboTag Strategy. (a) Colocalization of POMC (green) and HA (red) in ARC section of P21 Chow-O (white arrows). Images were taken at 20x (n = 3 biologically independent samples). (b) Average cell counting of exclusive POMC neurons (green bar), exclusive HA cells (red bars), and double POMC+ and HA + neurons (orange bars) (n = 3 biologically independent samples). (c) Enrichment expression analysis of specific neuropeptides (Pomc and Agrp) and negative control markers (Th and Aldh1L1) in POMC neuron-enriched mRNA samples from P21 Chow-O POMCRiboTag mice (n = 8 biologically independent samples). 3V: third ventricle. Data are expressed as mean ± SEM. Supplementary Figure 4Analysis of POMC neuron DEGs, belonging to neuronal generation GO categories, during P0 to P21 transition in Chow and HFD offspring. (a) Volcano plot illustrating DEGs of GO categories related to POMC neuronal generation (neurogenesis, generation of neurons, and neuron differentiation). (b) Average cell count and distribution throughout the ARC (bregma −1.06 to −2.46 mm) of POMC positive neurons in P21 Chow-O and HFD-O. n = 6–7 biologically independent samples. (c) BINGO analysis representing network relationships of biological processes based on DEGs belonging to GO categories related to POMC neuronal generation (neurogenesis, generation of neurons, and neuron differentiation). The circle size is proportional to number of genes that fall into a specific process, whereas the color intensity shows expression level differences. (d) CPM values of genes involved in POMC specification during P0–P21 transition in Chow and HFD offspring. Data are expressed as mean ± SEM. ∗p < 0.05. Supplementary Figure 5Analysis of POMC neuron DEGs, belonging to diverse GO categories, during P0 to P21 transition in Chow and HFD offspring. (a) Volcano plot illustrating DEGs of GO categories related to POMC neuronal maturation (nervous system development, system development, neuron development). (b) Volcano plot illustrating DEGs of GO categories related to POMC neuron projection development. DEGs are depicted in blue, red (upregulated), or green (common for Chow and HFD). The threshold for fold-change (±1.5) and FDR (<0.05) was considered. Unchanged genes are represented in black. N = 3–5 biologically independent samples. Supplementary Table 1List of the 227 differentially expressed TFs during POMC neuronal maturation identified by RNASeq (genes with a threshold for fold change (FC) of ±1.5 and false discovery rate (FDR) <0.05 are shown). For FC columns: white box (no change), blue box (downregulated), and pink box (upregulated). Green box in FDR columns indicates p < 0.05
References
- 1.Blüher M. Obesity: global epidemiology and pathogenesis. Nature Reviews Endocrinology. 2019;15(5):288–298. doi: 10.1038/s41574-019-0176-8. [DOI] [PubMed] [Google Scholar]
- 2.Friedman J.E. Developmental programming of obesity and diabetes in mouse, monkey, and man in 2018: where are we headed? Diabetes. 2018;67(11):2137–2151. doi: 10.2337/dbi17-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouret S.G. Development of hypothalamic circuits that control food intake and energy balance. In: Harris R.B.S., editor. Appetite and food intake: central control. 2nd ed. CRC Press/Taylor & Francis; Boca Raton (FL): 2017. [PubMed] [Google Scholar]
- 4.Toda C., Santoro A., Kim J.D., Diano S. POMC neurons: from birth to death. Annual Review of Physiology. 2017;79:209–236. doi: 10.1146/annurev-physiol-022516-034110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H., Simar D., Lambert K., Mercier J., Morris M.J. Maternal and postnatal overnutrition differentially impact appetite regulators and fuel metabolism. Endocrinology. 2008;149(11):5348–5356. doi: 10.1210/en.2008-0582. [DOI] [PubMed] [Google Scholar]
- 6.Fuente-Martín E., García-Cáceres C., Granado M., de Ceballos M.L., Sánchez-Garrido M.Á., Sarman B. Leptin regulates glutamate and glucose transporters in hypothalamic astrocytes. Journal of Clinical Investigation. 2012;122(11):3900–3913. doi: 10.1172/JCI64102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogt M.C., Paeger L., Hess S., Steculorum S.M., Awazawa M., Hampel B. Neonatal insulin action impairs hypothalamic neurocircuit formation in response to maternal high-fat feeding. Cell. 2014;156(3):495–509. doi: 10.1016/j.cell.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanz E., Yang L., Su T., Morris D.R., McKnight G.S., Amieux P.S. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(33):13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu A.W., Kaelin C.B., Takeda K., Akira S., Schwartz M.W., Barsh G.S. PI3K integrates the action of insulin and leptin on hypothalamic neurons. Journal of Clinical Investigation. 2005;115(4):951–958. doi: 10.1172/JCI24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culhane A.C., Thioulouse J., Perrière G., Higgins D.G. MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics (Oxford, England) 2005;21(11):2789–2790. doi: 10.1093/bioinformatics/bti394. [DOI] [PubMed] [Google Scholar]
- 13.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S. Orchestrating high-throughput genomic analysis with Bioconductor. Nature Methods. 2015;12(2):115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C., Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biology. 2001;2(8) doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wickham H. Springer; Dordrecht: 2009. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 17.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics (Oxford, England) 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 18.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouret S.G., Draper S.J., Simerly R.B. Trophic action of leptin on hypothalamic neurons that regulate feeding. Science (New York, N.Y.) 2004;304(5667):108–110. doi: 10.1126/science.1095004. [DOI] [PubMed] [Google Scholar]
- 20.Bouret S.G., Gorski J.N., Patterson C.M., Chen S., Levin B.E., Simerly R.B. Hypothalamic neural projections are permanently disrupted in diet-induced obese rats. Cell Metabolism. 2008;7(2):179–185. doi: 10.1016/j.cmet.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrey P., Maurin Y. Free-D: an integrated environment for three-dimensional reconstruction from serial sections. Journal of Neuroscience Methods. 2005;145(1–2):233–244. doi: 10.1016/j.jneumeth.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Biot E., Crowell E., Burguet J., Höfte H., Vernhettes S., Andrey P. Strategy and software for the statistical spatial analysis of 3D intracellular distributions. The Plant Journal: Cellular and Molecular Biology. 2016;87(2):230–242. doi: 10.1111/tpj.13189. [DOI] [PubMed] [Google Scholar]
- 23.Coupe B., Bouret S.G. Development of the hypothalamic melanocortin system. Frontiers in Endocrinology. 2013;4:38. doi: 10.3389/fendo.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padilla S.L., Carmody J.S., Zeltser L.M. Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nature Medicine. 2010;16(4):403–405. doi: 10.1038/nm.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanz E., Quintana A., Deem J.D., Steiner R.A., Palmiter R.D., McKnight G.S. Fertility-regulating Kiss1 neurons arise from hypothalamic POMC-expressing progenitors. Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2015;35(14):5549–5556. doi: 10.1523/JNEUROSCI.3614-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song A.J., Palmiter R.D. Detecting and avoiding problems when using the cre-lox system. Trends in Genetics: Trends in Genetics. 2018;34(5):333–340. doi: 10.1016/j.tig.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huisman C., Cho H., Brock O., Lim S.J., Youn S.M., Park Y. Single cell transcriptome analysis of developing arcuate nucleus neurons uncovers their key developmental regulators. Nature Communications. 2019;10(1):3696. doi: 10.1038/s41467-019-11667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry F.E., Sugino K., Tozer A., Branco T., Sternson S.M. Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. ELife. 2015;4 doi: 10.7554/eLife.09800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam B.Y.H., Cimino I., Polex-Wolf J., Nicole Kohnke S., Rimmington D., Iyemere V. Heterogeneity of hypothalamic pro-opiomelanocortin-expressing neurons revealed by single-cell RNA sequencing. Molecular Metabolism. 2017;6(5):383–392. doi: 10.1016/j.molmet.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimogori T., Lee D.A., Miranda-Angulo A., Yang Y., Wang H., Jiang L. A genomic atlas of mouse hypothalamic development. Nature Neuroscience. 2010;13(6):767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nasif S., de Souza F.S.J., González L.E., Yamashita M., Orquera D.P., Low M.J. Islet 1 specifies the identity of hypothalamic melanocortin neurons and is critical for normal food intake and adiposity in adulthood. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(15):E1861–E1870. doi: 10.1073/pnas.1500672112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jing E., Nillni E.A., Sanchez V.C., Stuart R.C., Good D.J. Deletion of the Nhlh2 transcription factor decreases the levels of the anorexigenic peptides alpha melanocyte-stimulating hormone and thyrotropin-releasing hormone and implicates prohormone convertases I and II in obesity. Endocrinology. 2004;145(4):1503–1513. doi: 10.1210/en.2003-0834. [DOI] [PubMed] [Google Scholar]
- 33.Aujla P.K., Naratadam G.T., Xu L., Raetzman L.T. Notch/Rbpjκ signaling regulates progenitor maintenance and differentiation of hypothalamic arcuate neurons. Development (Cambridge, England) 2013;140(17):3511–3521. doi: 10.1242/dev.098681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quarta C., Fisette A., Xu Y., Colldén G., Legutko B., Tseng Y.-T. Functional identity of hypothalamic melanocortin neurons depends on Tbx3. Nature Metabolism. 2019;1(2):222–235. doi: 10.1038/s42255-018-0028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarez-Bolado G. Development of neuroendocrine neurons in the mammalian hypothalamus. Cell and Tissue Research. 2019;375(1):23–39. doi: 10.1007/s00441-018-2859-1. [DOI] [PubMed] [Google Scholar]
- 36.Khodosevich K., Monyer H. Signaling in migrating neurons: from molecules to networks. Frontiers in Neuroscience. 2011;5:28. doi: 10.3389/fnins.2011.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell J.N., Macosko E.Z., Fenselau H., Pers T.H., Lyubetskaya A., Tenen D. A molecular census of arcuate hypothalamus and median eminence cell types. Nature Neuroscience. 2017;20(3):484–496. doi: 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu B., Shi X., Zhou Q., Chen H.S., Lim J., Han W. Hypothalamic NUCKS regulates peripheral glucose homoeostasis. Biochemical Journal. 2015;469(3):391–398. doi: 10.1042/BJ20150450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1HFD effects on maternal metabolism. (a) Maternal body weight before exposure to HFD (6 weeks of age), on the day of mating, and by the time of offspring's weaning (n = 16–24 biologically independent samples). (b-c) Maternal glucose tolerance test and area under the curve (AUC) at the onset of pregnancy (n = 8 biologically independent samples). (d) Maternal plasma insulin levels under fed and fasting conditions at the onset of pregnancy (n = 8 biologically independent samples). (e) Maternal gonadal white adipose tissue weight, relative to total body weight, in Chow and HFD females after weaning of their offspring (n = 21–29 biologically independent samples). Data are expressed as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001. Supplementary Figure 2Maternal HFD effects on female offspring metabolism. (a) Body weight at P21 in Chow and HFD female offspring (n = 6 biologically independent samples). (b-c) Glucose tolerance test and area under the curve (AUC) of P21 female offspring (n = 6 biologically independent samples). (d) Blood glucose levels at P21 of female HFD offspring after 6 h of food deprivation (n = 6 biologically independent samples). (e) Body weight in 12-week-old Chow-O and HFD-O females fed with a chow diet from weaning onward (n = 6–12 biologically independent samples). (f) Blood glucose levels, under fed and fasting conditions, of 12-week-old female HFD offspring (n = 6 biologically independent samples). (g-h) Glucose tolerance test and area under the curve (AUC) of 12-week-old Chow-O and HFD-O female offspring fed with a chow diet from weaning onward (n = 6 biologically independent samples). (i) Body weight of female Chow-O and HFD-O mice after 8 weeks of HFD exposure (n = 6 biologically independent samples). (j) Blood glucose levels under fed and fasting conditions after 8 weeks of HFD exposure in female Chow-O and HFD-O mice (n = 6 biologically independent samples). (k-l) Glucose tolerance test and area under the curve (AUC) after 8 weeks of HFD exposure in female Chow-O and HFD-O mice (n = 6 biologically independent samples). Data are expressed as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001. Supplementary Figure 3Validation of the RiboTag Strategy. (a) Colocalization of POMC (green) and HA (red) in ARC section of P21 Chow-O (white arrows). Images were taken at 20x (n = 3 biologically independent samples). (b) Average cell counting of exclusive POMC neurons (green bar), exclusive HA cells (red bars), and double POMC+ and HA + neurons (orange bars) (n = 3 biologically independent samples). (c) Enrichment expression analysis of specific neuropeptides (Pomc and Agrp) and negative control markers (Th and Aldh1L1) in POMC neuron-enriched mRNA samples from P21 Chow-O POMCRiboTag mice (n = 8 biologically independent samples). 3V: third ventricle. Data are expressed as mean ± SEM. Supplementary Figure 4Analysis of POMC neuron DEGs, belonging to neuronal generation GO categories, during P0 to P21 transition in Chow and HFD offspring. (a) Volcano plot illustrating DEGs of GO categories related to POMC neuronal generation (neurogenesis, generation of neurons, and neuron differentiation). (b) Average cell count and distribution throughout the ARC (bregma −1.06 to −2.46 mm) of POMC positive neurons in P21 Chow-O and HFD-O. n = 6–7 biologically independent samples. (c) BINGO analysis representing network relationships of biological processes based on DEGs belonging to GO categories related to POMC neuronal generation (neurogenesis, generation of neurons, and neuron differentiation). The circle size is proportional to number of genes that fall into a specific process, whereas the color intensity shows expression level differences. (d) CPM values of genes involved in POMC specification during P0–P21 transition in Chow and HFD offspring. Data are expressed as mean ± SEM. ∗p < 0.05. Supplementary Figure 5Analysis of POMC neuron DEGs, belonging to diverse GO categories, during P0 to P21 transition in Chow and HFD offspring. (a) Volcano plot illustrating DEGs of GO categories related to POMC neuronal maturation (nervous system development, system development, neuron development). (b) Volcano plot illustrating DEGs of GO categories related to POMC neuron projection development. DEGs are depicted in blue, red (upregulated), or green (common for Chow and HFD). The threshold for fold-change (±1.5) and FDR (<0.05) was considered. Unchanged genes are represented in black. N = 3–5 biologically independent samples. Supplementary Table 1List of the 227 differentially expressed TFs during POMC neuronal maturation identified by RNASeq (genes with a threshold for fold change (FC) of ±1.5 and false discovery rate (FDR) <0.05 are shown). For FC columns: white box (no change), blue box (downregulated), and pink box (upregulated). Green box in FDR columns indicates p < 0.05
Data Availability Statement
The RNASeq datasets generated and/or analyzed during the current study are available in the NCBI GEO repository under accession number GSE135639. [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135639].