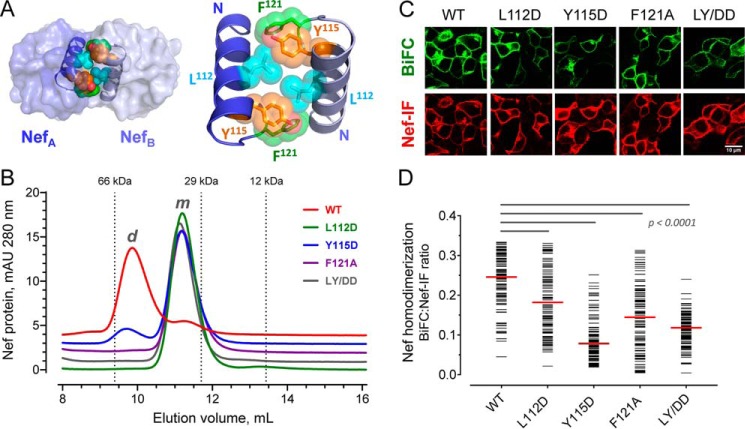
Figure 4.
Conserved hydrophobic residues in the folded Nef core are required for homodimer formation. A, Nef homodimers present in the X-ray crystal structure of the HIV-1 Nef core in complex with a Src-family kinase SH3 domain (PDB code 1EFN). An overview of the Nef dimer structure is shown on the left, with the αB helices that form the dimer interface highlighted; SH3 domains are not shown for clarity. The αB helices are enlarged on the right. Side chains of Leu112, Tyr115, and Phe121 from each Nef monomer form the hydrophobic core of the interface. B, analytical size-exclusion chromatography of WT Nef and dimerization interface mutants. WT Nef and the L112D, Y115D, F121A, and L112D/Y115D (LY/DD) mutants were expressed in E. coli and purified (see “Experimental procedures”). Each Nef protein was then characterized by analytical size-exclusion chromatography. Elution peaks of standard proteins are indicated by the vertical dotted lines and molecular weight. m, monomer; d, dimer. C, hydrophobic residues in the Nef core are required for homodimer formation in cells. WT Nef and the dimer interface mutants were expressed as BiFC pairs in 293T cells. The cells were stained for Nef expression with an anti-Nef antibody and imaged by confocal microscopy to detect Nef homodimer formation (BiFC, green) and Nef expression as immunofluorescence (Nef-IF, red). D, single-cell image analysis. Mean fluorescence intensities for the BiFC (interaction) and Nef-IF signals were determined with ImageJ for ≥100 cells. The Nef-BiFC:Nef-IF fluorescence intensity ratio for each cell is presented as a horizontal bar, with the median value indicated by the red bar. Student's t tests were performed on the groups indicated by horizontal lines above the plot; p < 0.0001 in each case.