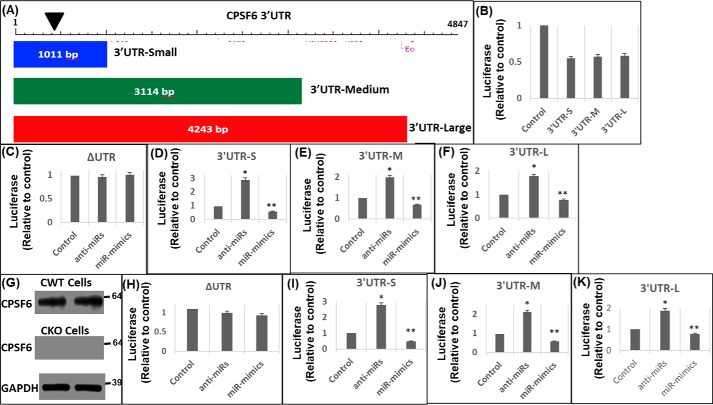
Figure 2.
miR-125b negatively regulates CPSF6 3′UTR activity. A, schematic representation of the three different sized DNA fragments of CPSF6 3′UTR that were cloned downstream of the luciferase stop codon to generate the corresponding reporter vectors: 3′UTR-small (3′UTR-S), 3′UTR-medium (3′UTR-M), and 3′UTR-large (3′UTR-L). The location of the miR-125b seed sequence is presented as an inverted black triangle. HEK-293T cells were transfected with either of these 3′UTR luciferase reporter vectors or the control pΔUTR (ΔUTR) vector in the presence or absence of miR-125b mimics or anti-miR-125b. 24 h post-transfection, luciferase activity was measured in the cellular lysates and plotted relative to the controls. B, luciferase activity of the CPSF6 3′UTRs and ΔUTR vectors. Luciferase activity of (C) ΔUTR, (D) 3′UTR-S, (E) 3′UTR-M, and (F) 3′UTR-L vector in the presence of anti-miRs or miR-mimics or scrambled controls. G–K, effects of altered miR-125b levels on CPSF6 3′UTR activity in CKO cells. Immunoblots showing CPSF6 expression in WT HEK-293T (CWT) (G, top panel) and CKO cells (G, middle panel), and GAPDH (loading control) expression in CKO cells (G, bottom panel). Luciferase activity in CKO cells transfected with (H) ΔUTR, (I) 3′UTR-S, (J) 3′UTR-M, and (K) 3′UTR-L vector in the presence of anti-miRs or miR-mimics or scrambled controls in CKO cells. Error bars represent S.E., whereas, *, p < 0.05 represents the comparison of anti-miR or miR mimic samples versus scrambled controls.