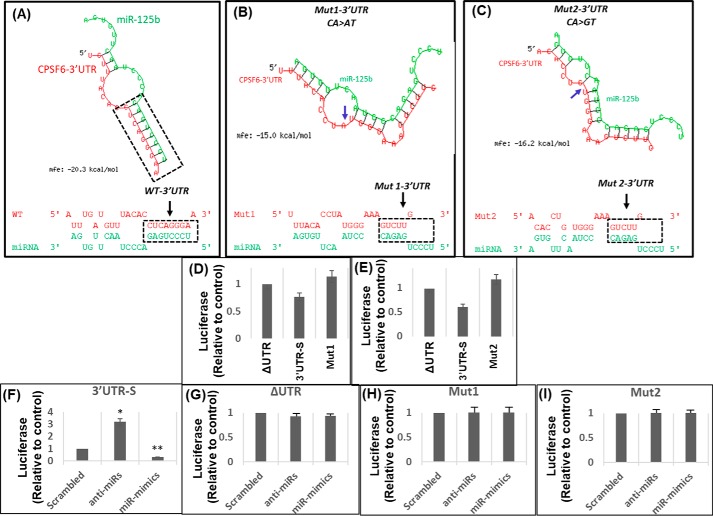
Figure 3.
Mutations in the miR binding sequence abrogate the regulatory effects of miR-125b on the CPSF6 3′UTR. A–C, in silico analysis of the effects of two different mutations, Mut1 (CA > AT) and Mut2 (CA > GT), introduced at the miR-125b–binding site in CPSF6 3′UTR on their binding stability and affinity (as measured by mfe) and the secondary hairpin loop structure integrity. Shown are the effects of Mut1 (B) and Mut2 (C) compared with WT CPSF6 3′UTR (A). D and E, effects of endogenous miR-125b on the activity of the CPSF6 3′UTR mutants, Mut1 and Mut2. Shown is the comparison between the luciferase activity in the lysates of HEK-293T cells transfected with Mut1 (D) or Mut2 (E) and the luciferase activity of ΔUTR and 3′UTR-S. F–I, effects of miR-125b overexpression or down-regulation on the activity of the CPSF6 3′UTR mutants, Mut1 and Mut2. Luciferase activity in lysates prepared from HEK-293T cells transfected with (F) 3′UTR-S, (G) ΔUTR, (H) Mut1, or (I) Mut2 reporter constructs in the presence or absence of anti-miRs or miR-mimics or scrambled controls. Data presented are mean values of three independent experiments with error bars representing S.E. *, represents p < 0.05 for the comparison of miR-mimics or anti-miR versus scrambled controls.