Figure 5. Diabetes-associated regions form hyperconnected 3D cliques in NOD mice.
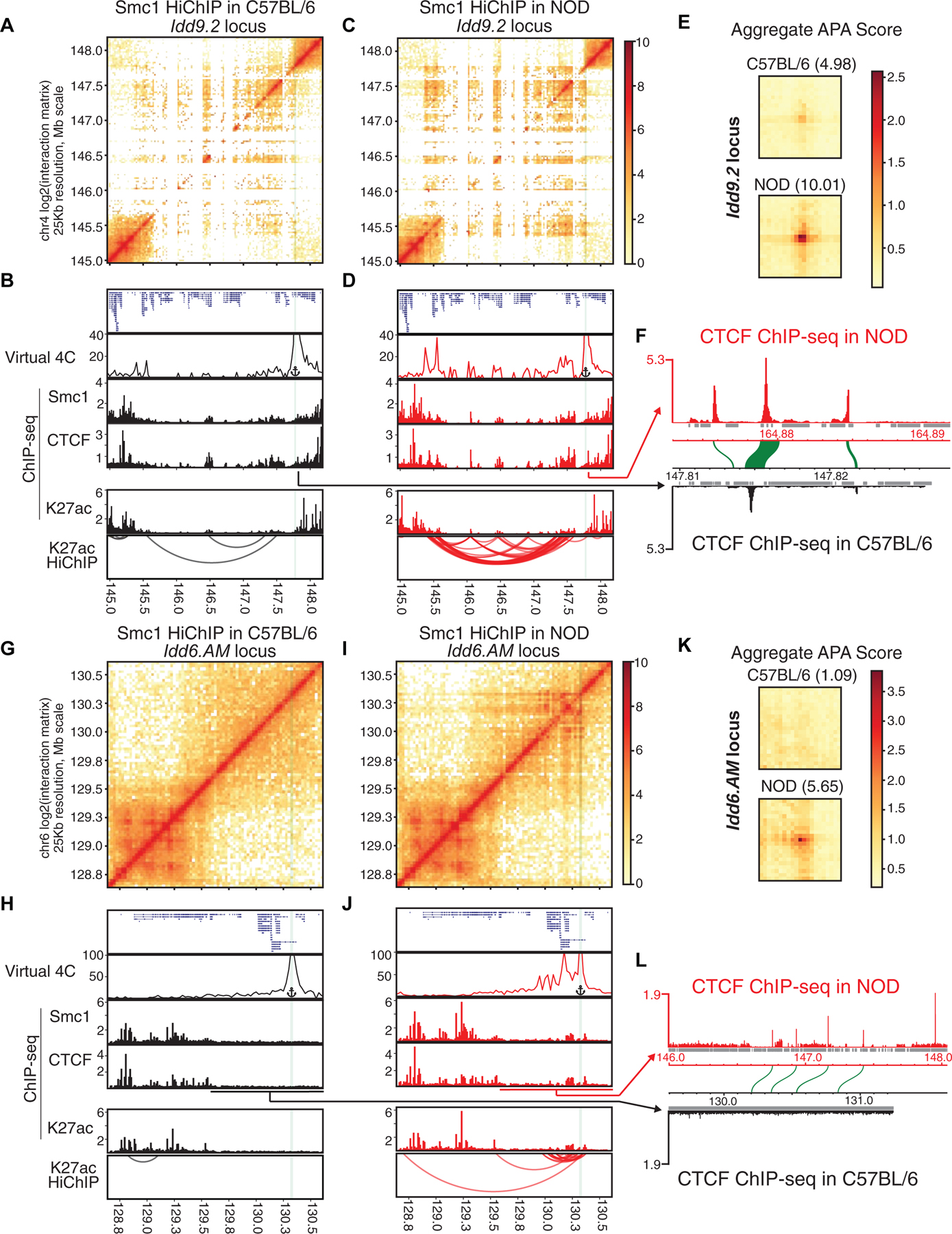
(A-L) Contact matrices based on Smc1 HiChIP in C57BL/6 and NOD strains at the NOD-specific hyperconnected regions harboring Idd9.2 (A, C) and Idd6.AM (G, I). Virtual 4C plot at the highlighted anchor, genome browser views of Smc1, CTCF, H3K27ac ChIP-seq and reproducible significant loops from H3K27ac HiChIP for Idd9.2 (B, D) and Idd6.AM (H, J) in both strains. Virtual 4C plot quantifies the connections of the highlighted anchor to other regions. (E, K) Aggregate peak analysis (APA) was used to measure the strength of the Smc1 HiChIP loops in contact matrices from both strains in Idd9.2 (E) and Idd6.AM regions (K).
(F and L) CTCF binding in NOD de novo assembled genome and the orthologous relationship with C57BL/6 genome in the syntenic regions of Idd9.2 (F) and Idd6.AM (L). At the Idd9.2 locus in the NOD genome (F), we observed a region at the boundary containing three CTCF binding events, of which the sequence of only one binding site is well conserved in C57BL/6 genome (synteny of CTCF binding events are depicted as green arcs). At the Idd6.AM locus (L), there is one unique CTCF binding site in NOD genome that does not have orthologous sequence in the corresponding region in C57BL/6 genome (L). Repetitive regions are depicted in grey.
