Figure 3.
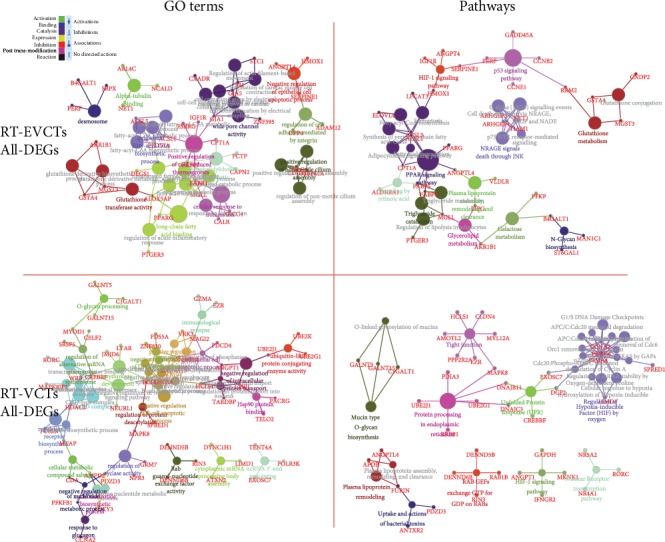
GO and pathway terms associated with all DEGs in RT-EVCTs and RT-VCTs. All DEGs were submitted separately according to their cell type of origin to ClueGO with the default parameters. GO and pathway enrichment were set up for analysis. DEGs were classified by three ways: by GO biological process, GO molecular function, and GO cellular component. The KEGG and Reactome database was consulted to determine pathway enrichment. An exhaustive list of all terms (including those not shown above) can be found in supplementary materials (Tables ). DEGs: differentially expressed genes; RT-EVCTs: rosiglitazone-treated extravillous cytotrophoblasts; RT-VCTs: rosiglitazone-treated villous cytotrophoblasts; GO: gene ontology; KEGG: Kyoto encyclopedia of genes and genomes.
