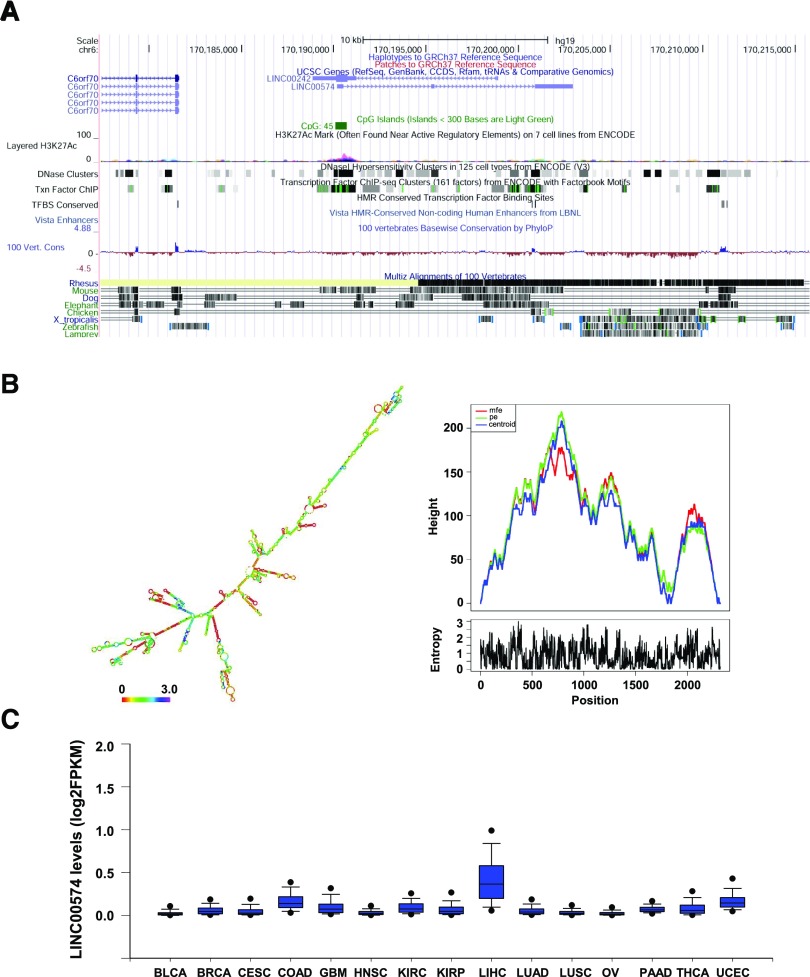
Fig. 2.
LINC00574 is a liver-specific lncRNA with a stable secondary structure. (A) Schematic representation of LINC00574 on chromosome 6 based on University of California Santa Cruz Genome Browser tracks showing CpG island information, peak signal of H3K27Ac, mammalian conservation. (B) Secondary structure (color indicates positional entropy) and mountain plot of LINC00574. A secondary structure of LINC00574 was predicted based on minimal free energy (–1060.10 kcal/mol, left panel). Right panel shows a mountain plot of minimum free energy structure, centroid structures, and partition function structure. The closely related status for these curves indicated the stable secondary structure of RNA molecule. The height (y-axis) of the mountain plot indicates the number of base pairs enclosing a sequence position indicated in the x-axis. pe indicates partition function structure; Centroid indicates centroid structure. (C) RNA levels (log2 fragments per kilobase of transcript per million mapped reads) of LINC00574 in 15 different types of tissues (tumors and adjacent normal tissues) from TCGA data base. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, colon adenocarcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma.