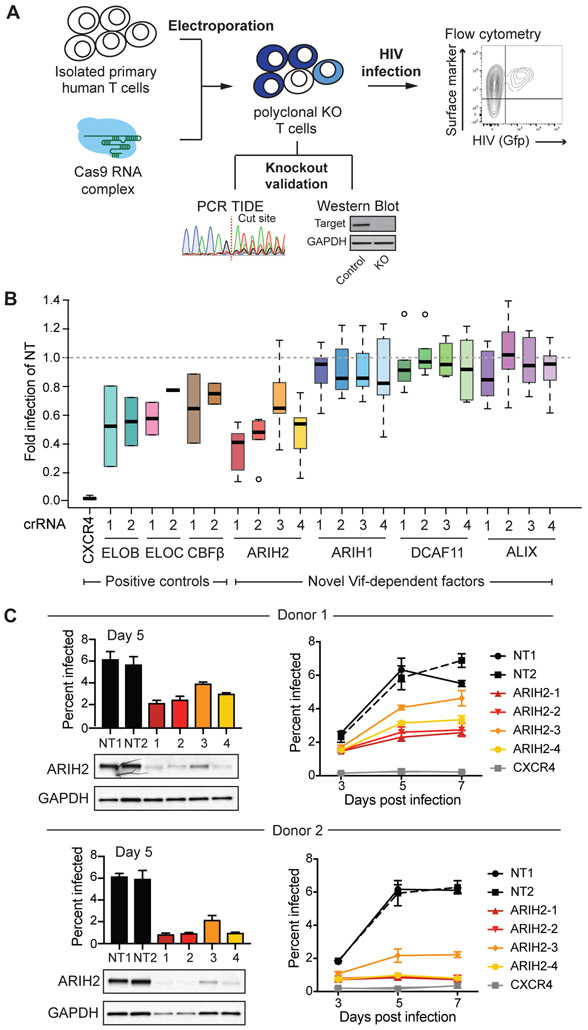
Figure 4. ARIH2 knockout in primary T cells leads to a reduced HIV infection rate.
(A) Workflow for functional validation of CUL5 factors using CRISPR knockouts followed by HIV infection in primary T cells, adapted from Hultquist et al (Hultquist et al., 2016).
(B) Relative HIV infection rate in primary T cells comparing CRISPR knockouts of selected genes to non-targeting control (day 5 of spread infection, NT = non-targeting control). Each gene was knocked out using 2-4 guide RNAs in 2-6 donors, infections for each donor and knockout were performed in triplicates.
(C) Western blots confirming ARIH2 knockout for all four guide RNAs in primary T cells from two donors and the corresponding HIV infection rate at day 5 of spread infection. For each donor spread HIV infections rate over seven days is shown for each ARIH2 guide RNA compared to controls, non-targeting guide RNA and CXCR4 knockout (n = 3, data from three replicate infections are presented as mean ± SEM, NT = non-targeting control).
See also Figure S2.