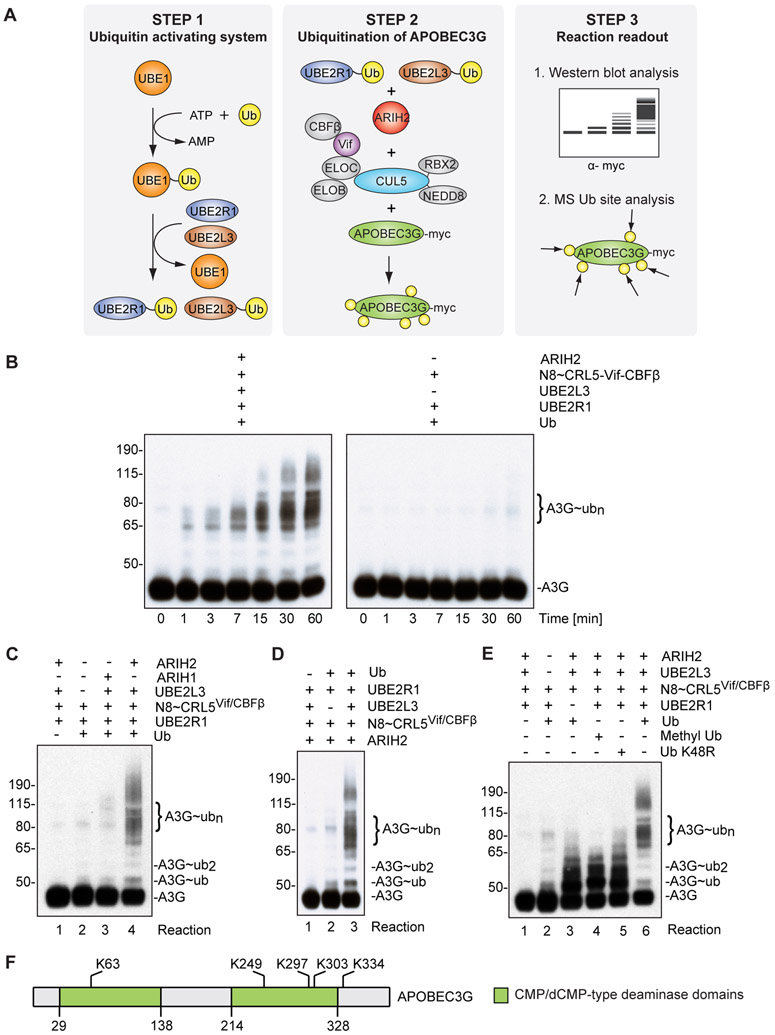
Figure 5. ARIH2 monoubiquitinates APOBEC3G to prime it for accelerated polyubiquitination via CUL5Vif/CBFß in vitro.
(A) Schematic describing the in vitro ubiquitination reaction.
(B) Immunoblot showing time course for APOBEC3G ubiquitination reactions in the presence and absence of ARIH2/UBE2L3. In the presence of ARIH2 and UBE2L3, formation of extensive ubiquitination chains on APOBEC3G via neddylated CUL5vif/CBFß (N8~CRL5Vif/CBFß) is accelerated and enhanced compared to treatment with neddylated CUL5vif/CBFß and UBE2R1 alone.
(C) Acceleration of APOBEC3G ubiquitination is specific for ARIH2 and cannot be phenocopied by replacing ARIH2 with ARIH1 (Reaction 3).
(D) Acceleration of APOBEC3G ubiquitination is dependent on UBE2L3.
(E) ARIH2 monoubiquitinates A3G at multiple sites to prime for CRL5Vif/CBFß dependent Ub chain extension. Immunoblot showing APOBEC3G ubiquitination reactions in the presence and absence of UBE2R1, ARIH2 and CUL5vif/CBFß, Ub K48R, Methyl-Ub or wild-type Ub. Methyl-Ub (Reaction 4) and K48R-Ub (Reaction 5) recapitulate the pattern observed for the reaction that focuses on ARIH2 activity on A3G ubiquitination (Reaction 3, absence of UBE2R1).
(F) Ubiquitination sites on APOBEC3G derived from the in vitro assay in the presence of ARIH2 and UBE2L3 overlap with previously described APOBEC3G sites in cells (Albin et al., 2013).
See also Figure S3.