Abstract
Large clinical trials and studies in model systems suggest that the chemical form of selenium may dictate chemopreventive and chemotherapeutic efficacy. Selenite induces excess ROS production, which mediates autophagy and eventual cell death in non-small cell lung cancer (NSCLC) adenocarcinoma A549 cells. The exact mechanisms that underlie these phenotypic effects are unclear, thus the clinical relevance of selenite or other selenocompounds for cancer therapy remains to be determined. Using stable isotope-resolved metabolomics (SIRM) and gene expression analysis, we previously showed that selenite simultaneously disrupts glycolysis, the Krebs cycle, and polyamine metabolism in A549 cells, potentially through perturbed glutaminolysis. Glutaminolysis is a vital anaplerotic process for the proliferation and survival of many cancer cells. Here we investigated the role of key glutaminolytic enzyme glutaminase 1 (GLS1) in selenite’s toxicity in A549 cells and in patient-derived lung cancer tissue slices. Using [13C6]-glucose and [13C5,15N2]-glutamine as tracers, we tracked the time course of selenite’s action on multiple pathways. We found that selenite inhibited glutaminolysis and glutathione synthesis by suppressing GLS1 expression in addition to blocking the Krebs cycle but transiently activating the pyruvate carboxylase activity. Glutamate supplementation in part rescued these anti-proliferative and oxidative stress activities. Similar metabolic perturbations and tissue necrosis were evident in human patients’ cancerous lung tissues in ex vivo experiments with selenite. Altogether, our results support the hypothesis that GLS1 suppression mediates part of the anti-cancer activity of selenite both in vitro and ex vivo.
Keywords: lung cancer; selenite; glutaminolysis; stable isotope resolved metabolomics; [13C6]-glucose; [13C5,15N2]-glutamine
Introduction
Selenium is a micronutrient element that is incorporated into the 21st protein-coding amino acid, selenocysteine[1]. It functions primarily as an anti-oxidant via its catalytic role in redox enzymes such as thioredoxin reductase and glutathione peroxidase[2]. A selenium deficient diet leads to a constellation of syndromes including bone deformities, thyroid disorders, weakened immune response, and an endemic cardiomyopathy of the Keshan province in China[3]. Selenium agents have also been shown to display anti-cancer and chemopreventive activity in several large-scale clinical trials[4–6]. As with any element, the anti-cancer or chemopreventive potential of selenium is highly dependent on the chemical form that is used, and a wide variety of organic and inorganic compounds have been studied. The discrepancy in efficacy of the various selenocompounds could have been responsible for the failure of the large SELECT trial (which used pure selenomethionine and vitamin E)[4], and thus further research into the toxicity of different Se agents in human cancer cells and tissues is needed.
Selenite (SeO32-) is one of the main naturally occurring inorganic forms of selenium that has promising chemopreventive and chemotherapeutic effects [5]. Selenite has been shown to induce dose-dependent apoptosis in a variety of cancer cells including lung, prostate, glioma, and colorectal cancers [7, 8]. Park et al. [9] uncovered evidence of reactive oxygen species (ROS)-mediated autophagy in A549 lung adenocarcinoma cells eventually leading to cell death. However, despite these findings, the exact mechanism of selenite-induced toxicity in cancer cells remains unclear. One of the main theories involves the generation of ROS such as superoxide, hydrogen peroxide, and/or hydroxyl radical from the successive reduction of SeO32- to elemental selenium (Se0) [10]. The ROS generated can then induce DNA and protein damage that precipitate a cascade of events eventually leading to apoptosis and cell death. ROS accumulation and cell death are blocked in selenite-treated A549 cells by supplementation with N-acetylcysteine (NAC), a commonly used antioxidant, which supports this hypothesis [9].
We have previously used [13C6]-glucose tracing coupled with metabolomics (i.e. stable isotope-resolved metabolomics, or SIRM) to show that selenite induced numerous perturbations of metabolic pathways in A549 cells including disruption of the Krebs cycle, urea cycle, polyamine metabolism, and glycolysis. Altered glutaminolysis was also implicated based on gene expression data [11]. The utility of SIRM has also been demonstrated in elucidating metabolic reprogramming in human lung cancer tissues in vivo [12, 13], which was recapitulated in ex vivo tissue slice cultures [13–15]. The patient-derived (PD) ex vivo tissue models have the advantages of maintaining human tumor 3D architecture and microenvironment while enabling comparison of matched cancerous (CA) versus non-cancerous (NC) tissues, and flexible experimental designs including use of tracers and treatment agents.
Glutamine (Gln) is a major anaplerotic substrate of the Krebs cycle and contributes to the synthesis of nucleotides, amino acids, hexosamines, and glutathione among other metabolites. Gln consumption is often increased in tumors and the inhibition of the key catabolic enzyme glutaminase (GLS) has been shown to induce apoptosis in kidney and lung cancer cells overexpressing MYC, the proto-oncogene that drives GLS expression[16]. We and others have found that c-MYC expression is elevated in >50% of non-small cell lung cancer (NSCLC) tumors relative to adjacent benign tissue in patient studies [17]. Recently it has been shown that selenite inhibits glutaminolysis by inducing degradation of GLS1 in colorectal cancer [8]. However, a direct evaluation of the downstream metabolic consequences of selenite inhibition of glutaminase, and whether there are other targets, has not been reported.
Based on our previous findings on disrupted Gln metabolism-related gene expression by selenite in A549 cells[18] and the large body of literature surrounding the importance of Gln for tumor nutrition, we investigated Gln metabolism with a focus on GLS as a possible mechanism for selenite-induced toxicity in human NSCLC. We employed [13C5,15N2]-Gln and SIRM to track changes in Gln metabolism induced by selenite in two human NSCLC models, i.e. A549 cell and ex vivo PD tissue slice cultures. We found that selenite blocked glutaminolysis by suppressing GLS1 expression, thereby disrupting glutamine to glutamate (Glu) conversion and glutathione metabolism while inducing ROS and cell death in both A549 cell and ex vivo tissue models. This toxic mechanism was supported by Glu rescue of selenite-induced growth inhibition and glutathione depletion in A549 cells.
Materials and Methods
Cell culture treatments:
A549 cells were grown in glucose-free, Gln-free DMEM medium supplemented with 10% FBS, 0.2% glucose (w/v), 4 mM glutamine, and 100 I.U./mL penicillin 100 μg/mL streptomycin at 37 °C and 5% CO2. Na2SeO3 solutions were prepared immediately prior to use and sterilized using a 0.22 μm syringe filter. For time course tracer experiments, cells were incubated with half volume of the tracer medium for two hours prior to treatment in order to condition cells with the tracer medium before treatment. The remaining volume of media containing 2x Na2SeO3 was then added. For CB-839 treatments, A549 cells were grown in the above DMEM medium with Gln replaced by [13C515N2]-Gln plus 1 μM CB-839 (maximal solubility in media) or vehicle (DMSO) for 24, 48, or 72 h. For Glu supplementation experiments, A549 cells were grown in the above DMEM medium ± 6.25 μM Na2SeO3 ± 5 mM [13C515N]-Glu for 24 h.
shRNA –
Lentivirus was produced in HEK293T cells using psPAX2 and pMD2.G packaging and envelope vectors (gifts from Didier Trono; Addgene plasmids #12260 #12259, respectively)) and pLKO.1-based transfer vectors (Sigma Aldrich). A549 cells were transduced with lentivirus plus 8 μg/mL polybrene for 24 h. Cells were split to allow for clonal expansion 48 h later. Then, 1 μg/mL puromycin was added to select for stable integrants. Cells remained in the selection medium until sufficient cells had grown for experimental needs. Targeting sequences - shGLS1–1 (clone ID NM_014905.2–1441s1c1): 5’ – GCACAGACATGGTTGGTATAT −3’; shGLS1–2 (clone ID NM_014905.2–1576s1c1): 5’- GCCCTGAAGCAGTTCGAAATA −3’. Non-targeting control was catalog #SHC016 from Sigma Aldrich.
Fresh tissue slice treatments:
During NSCLC surgical resection, very thin slices (0.5–1 mm) were freshly prepared from CA and adjacent NC tissues by the operating surgeon (Dr. M. Bousamra II). Tissue slices were immediately placed in T-25 culture flasks with 8 mL DMEM medium supplemented with 10% FBS, 100 I.U./mL penicillin 100 μg/mL streptomycin, 0.2% glucose, and 4 mM Gln. For tracer experiments, [13C6]-glucose or [13C515N2]-Gln replaced the unlabeled counterparts, as described previously[15, 19] with the addition of 6.25 μM Na2SeO3 or 1 μM CB-839 to the DMEM medium. Parallel treatments with the corresponding vehicles (PBS for Na2SeO3 and DMSO for CB-839) were also performed. Flasks were then placed on a rotator in a cell culture incubator at 37 °C and 5% CO2 for 24 h. After 24 h, media were collected and tissue slices were washed with PBS before flash freezing in Liq. N2 for downstream processing as described below.
Metabolite quenching and extraction:
At the time of A549 cell harvest, media were collected then all plates were quickly washed three times with ice cold PBS, and PBS was removed by aspiration. Cell metabolism was quenched with 1 mL of −20 °C CH3CN before adding 0.75 mL of Nano pure water and harvesting by scraping. The process was repeated once before CHCl3 addition to a final CH3CN:H2O:CHCl3 ratio of 2:1.5:1 to obtain the polar, non-polar and insoluble proteinaceous interface fractions for downstream analysis [20]. For extraction of metabolites from human tissue slices, frozen tissue was first pulverized to powder in a liquid nitrogen freezer mill (SPEX SamplePrep, Metuchen, NJ). The frozen powder was then extracted using the same ratio of CH3CN:H2O:CHCl3 described above.
Protein hydrolysis:
The residues from the CH3CN:H2O:CHCl3 extraction were extracted for proteins with 62.5 mM Tris + 2% sodium dodecyl sulfate (SDS) + 1 mM dithiothreitol (DTT) at pH 6.8. The proteins were precipitated, washed, and hydrolyzed to individual amino acids with 6N HCl using a focused-beam microwave as described previously [21]. The hydrolysates were lyophilized before derivatization with MTBSTFA and GC-MS analysis as described below.
NMR experiments:
NMR experiments were conducted on a Varian Inova 14.1 T instrument equipped with 5 mm HCN cold probe or an Agilent DD2 spectrometer equipped with a 3 mm HCN cold probe. Cell extracts were dissolved in D2O with 30 nmol d6-DSS, as internal standard and loaded into 5 mm Shigemi or 1.7 mm NMR tubes. 1D proton spectra were recorded with an acquisition time of 2 seconds after initial presaturation of the H2O signal with a recycle time of 6 seconds. 1D 13C-filtered heteronuclear single quantum coherence (HSQC) spectra were measured using an acquisition time of 150 ms and a recycle time of 2 s. 1D proton spectra were processed using a 1 Hz exponential line broadening function and a zero fill of 128 k points. 1D 13C-filtered HSQC spectra were processed with a 4 or 6 Hz line-broadening exponential and an unshifted Gaussian function and zero filled to 16 k points. Both types of spectra were processed manually for baseline and phase corrections. 1D proton spectra were referenced using the methyl signal of d6-DSS at 0 ppm, and 1D 13C-filtered HSQC spectra were referenced to the C-4-Gln peak at 2.43 ppm. Metabolites were identified in reference to an in-house standard database and quantified by calibration with the methyl resonance of the DSS internal standard using the MestReNova NMR processing software (Mestrelab Research, Santiago de Compostela, Spain)[22].
GC-MS:
Briefly, an aliquot of the lyophilized extracts was derivatized in 50% N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-Butyldimethylchlorosilane (MTBSTFA + 1% TBDMSCl) in CH3CN before GC-MS analysis. All GC-MS data were acquired on a ThermoFinnigan PolarisQ GC-Ion trap MSn instrument (ThermoScientific, Rockford IL). One μL of the derivatized sample was separated on a SGE Forte (Victoria, Australia) fused silica open-tubular column with the following dimensions: 50 m x 0.15 mm; 0.25 μm film thickness of 5% phenylmethyl-phenylsiloxane. The GC parameters were as follows: injector and transfer line, 280 °C; oven, 60 °C for 2 min, 60 to 150 °C at 20 °C/min ramp, 150 to 300 °C at 6 °C/min ramp; He carrier gas with flow rate of 1.5 mL/min. Spectra were acquired in segmented scan mode with ranges of 140–206, 209–280, and 283–650 m/z; all scans were completed at a rate of 5 spectra/sec which were averaged into one. Spectra were processed using Xcalibur Quan Browser MS software and quantified using a mixture of 63 standards at 50 μM each except for glutamate at 500 μM. All samples and standards were spiked with 1 μM norleucine as internal standard.
Ion chromatography-ultra high-resolution Fourier transform-MS (IC-UHR FT-MS) was performed as previously described [15]. Briefly, polar fractions were reconstituted in 20 μL Nanopure water, and analyzed by a Dionex ICS-5000+ ion chromatograph interfaced to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) operating at a resolution setting of 500,000 (FWHM at m/z 200) on MS1 acquisition to detect any and all combinations of 13C and 15N isotopologues. The chromatograph was outfitted with a Dionex IonPac AG11-HC-4 μm RFIC&HPIC (2 × 50 mm) guard column upstream of a Dionex IonPac AS11-HC-4 μm RFIC&HPIC (2 × 250 mm) column. Chromatography and mass spectrometric settings were the same as described previously [15] with an m/z range of 80 to 700. Peak areas were integrated and exported to Excel via the TraceFinder 3.3 (Thermo) software package. Peak areas were corrected for natural abundance as previously described[23], after which fractional enrichment and μmoles metabolites/g protein were calculated to quantify 13C incorporation into various pathways.
Western Blotting:
At time of harvest cells were lysed directly on the plate using 62.5 mM Tris, 2% SDS buffer at pH 6.8 before harvest by scraping, and protein concentration was measured via the BCA method (Pierce). Approximately 10 μg protein was loaded for each sample onto a denaturing SDS 10 % acrylamide gel, electrophoresed, and transferred to nitrocellulose membrane. All primary antibodies were incubated with membranes at 4 °C on a rocker overnight at appropriate dilutions in PBST + 5% non-fat powdered milk as follows: GLS1 (GeneTex, GTX81012), 1:1000; PCB, 1:500 (Santa Cruz, H-300); α-tubulin 1:50,000 (Epitomics, 1878–1); β-actin (Sigma, A5441) 1:10,000. Membranes were then washed 3 times with PBS + 0.1% Tween-20 (PBST) before incubating 1 hour at room temperature with an HRP-conjugated secondary antibody (Thermo Fisher, 1:10000 in PBST with 5% non-fat powdered milk). Blots were then incubated in Pierce SuperSignal Dura chemiluminescent reagent before documentation using X-ray film or a ChemiDoc-It system (UVP). Expression was quantified using ImageJ software.
qRT-PCR:
At the time of harvest, total RNA was extracted from cells using Trizol reagent (Invitrogen) and cDNA was synthesized using Invitrogen Superscript VILO cDNA synthesis kit. qRT-PCR was performed using Maxima SYBR green based mastermixes (Thermo Fisher) according to the manufacturer’s instructions. qRT-PCR was quantitated using the ΔΔCt method with 18 S rRNA used as the reference gene. GLS1 primer pair 1: forward – 5’ CACATTGATGAGTTATATGAAAGTGC- 3’ and reverse – 5’ GGCCAGTTGAGGAATATAATCTG 3’. GLS1 primer pair 2: forward – 5’ GTGCCGTTATTTACCCCTGA 3’ and reverse – 5’ TTACCAGCCAACAGCAAACA 3’. 18S: forward - 5’ AACGGCTACCACATCCAA 3’ and reverse – 5’ GACTCATTCCAATTACAGGGC 3’.
Microscopy:
A549 cells were grown to ~70% confluence in 6-well plates containing sterilized cover slips before treating with 6.25 μM Na2SeO3. At 3, 6, 24 and 48 hours medium was aspirated. For ROS measurement 10 μM H2-DCFDA (Molecular Probes) in fresh medium was added to each well and incubated at room temperature for 20 min. Medium was then replaced with fresh medium and cells were incubated for another 30 min at room temperature to allow dye to hydrolyze. For microscopy, cells in multi-well chamber slides were washed in PBS before adding cover slips and the edges were sealed with fingernail polish. For microplate reading, cells in 96-well plates were analyzed for fluorescence with excitation and emission wavelengths at 492 and 527 nm, respectively. For mitochondrial staining, 25–100 nM MitoTracker Red FM (Molecular Probes) in fresh medium was added to each well in multi-well chamber slides and incubated at 37 °C for 0.5–1 hour as described by the manufacturer. The slides were processed and sealed as described above. Images were recorded on an Olympus BX51 fluorescence microscope and appropriate filters (Omega Optical, Brattleboro, VT).
Proliferation assays:
A549 cells were grown in 6-well plates and fixed with 70% methanol for 30 minutes at −20° C, rinsed twice with 1x PBS, then stained with DAPI for 10 minutes at room temperature. DAPI stain was removed and cells again rinsed twice with 1xPBS. Cells were imaged on a Cytation 3 plate reader (BioTek) and counted using the Gen5 software.
GLS Activity assays.
Phosphate-dependent glutaminase activity was measured in mitochondrial fractions by the rate of ammonia formation as described previously [24].
Statistics:
All metabolite amounts were normalized to total protein measured via the BCA method. All error bars represent the standard error of the mean. For statistical significance of experiments where only a single variable was measured the two-tailed Student unpaired t-test was used to calculate p values; p values <0.05 were considered significant. For experiments involving multiple measurements (i.e., measurement of metabolites from NMR and MS analysis) the false discovery rate method described by Benjamini and Hochberg was applied [25]. Tests with p values <0.05 and an FDR q value <0.05 were then accepted as significant. For cell-based experiments, only Glu, Gln, and oxidized/reduced glutathione (GSSH/GSH) were measured for statistical differences among treatments. For tissue experiments Glu, fumarate, malate, and aspartate (Asp) were measured.
Results
Selenite disrupts Gln utilization in human lung cancer models
A. Cell culture model
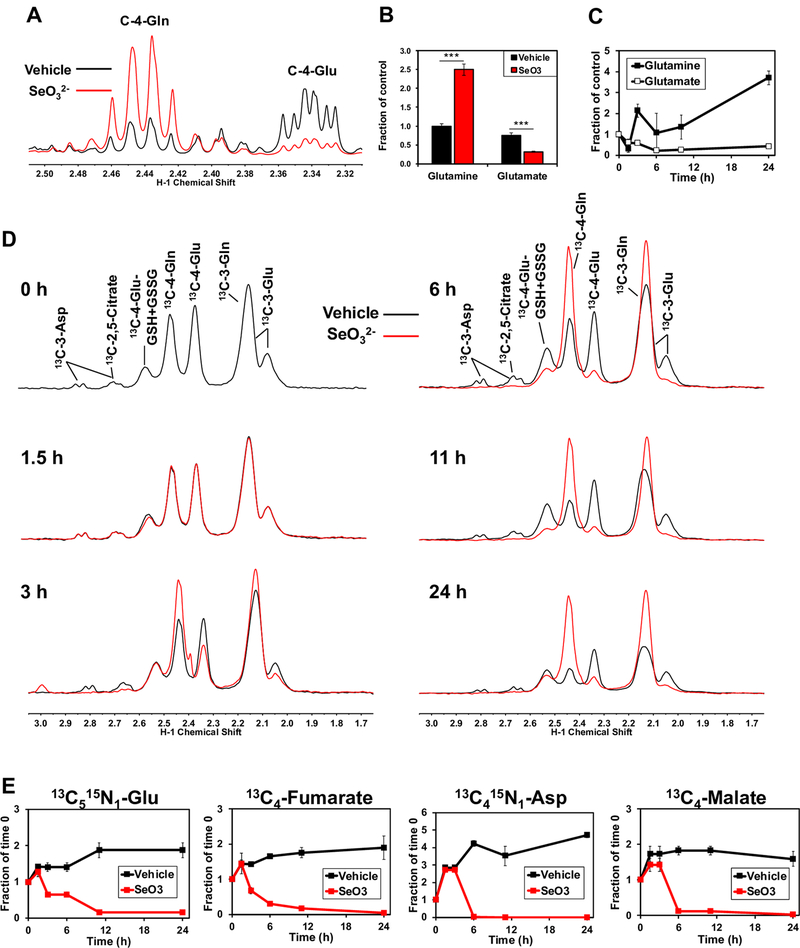
To determine the metabolic effects of SeO32-, we incubated A549 human lung cancer cells with 6.25 μM Na2SeO3, which was previously established as the IC50 for A549 cell growth at 24 h [26]. Metabolites in polar extracts were then analyzed using NMR and GC-MS as described in the Methods. A549 cells treated with selenite showed Gln accumulation and Glu depletion (Fig. 1A) with the Gln level elevated about 2.5-fold and Glu level decreased by 3-fold (Fig. 1B). Time-course measurements by GC-MS of selenite treated A549 cells revealed that these changes in Gln and Glu levels began at 3 h post-treatment and continued for the duration of the experiment (Fig. 1C). By 6 h of selenite treatment, Glu depletion had plateaued with a half-life of approximately 2 h, while Gln continued to accumulate suggesting that selenite disrupts glutamine hydrolysis without affecting its uptake.
Figure 1. Selenite impairs glutaminolysis and blocks the synthesis of downstream metabolites in A549 cells.
A) 1D 1H NMR spectra were recorded from polar extracts of A549 cells treated for 24 h with 6.25 μM Na2SeO3. Spectra were superimposed and normalized to total protein to assess differences between treatments in the intensities of the C-4-Gln and C-4-Glu peaks at 2.44 and 2.34 ppm, respectively. B) The C4-Glu and Gln peaks were quantified and normalized to total protein. ***q<0.001, ** q<0.01. C) Total Gln and Glu normalized to total protein were obtained from GC-MS analysis of polar extracts recorded at 0, 1.5, 3, 6, and 24 h of selenite treatment. D) 1D 1H{13C} HSQC spectra were recorded from polar extracts of A549 cells pre-incubated for 2 hours with 13C5,15N2-Gln tracer and treated with or without 6.25 μM Na2SeO3 for 0, 1.5, 3, 6, 11, and 24 hours. Control and selenite spectra were superimposed and normalized to total protein to visualize differences between treatments in 13C-4-glutathione, 13C-4-Gln, and 13C-4-Glu at 2.54, 2.44, and 2.34 ppm, respectively. E) GC-MS data were acquired from these extracts and 13C5,15N1-Glu (m+6), 13C4-malate, 13C4-fumarate, and 13C4,15N1-Asp (m+5) were quantified and normalized to total protein.
To confirm the selenite effect on glutaminolysis and to follow the fate of the product Glu in cell experiments, we treated A549 cells with selenite using [13C5,15N2]-Gln as the tracer to directly measure the uptake of Gln and its conversion to Glu and downstream metabolites, using 1D 1H{13C} HSQC NMR. Consistent with findings from unlabeled metabolite profiling, the selenite-induced [13C]-Gln buildup and [13C]-Glu depletion began at 3 h and continued thereafter (Fig. 1D). Also shown in Fig. 1D is the depletion of 13C-labeled Asp and citrate beginning at ~3 h of selenite treatment. Furthermore, [13C515N2]-Gln derived products of glutaminolysis and the Krebs cycle including [13C5,15N1]-Glu, [13C4]-fumarate, [13C4]-malate, and [13C4,15N1]-Asp showed a progressive decrease with selenite treatment, which began at 3 h to nearly undetectable biosynthesis by 24 h (Fig. 1E). In contrast, the corresponding metabolites in the controls continued to increase for the duration of the experiment, possibly because the Krebs cycle had not yet reached isotopic steady state.
We also followed selenite-induced time course changes in the metabolism of [13C6]-glucose in A549 cells. Selenite treatment resulted in large decreases in the levels of 13C2-citrate (A), -malate (C), and -Glu (E) as well as 13C3-Glu (F) (■ versus ■) 24 h (Fig. S1). This is consistent with attenuation in the pyruvate dehydrogenase (● PDH)-initiated Krebs cycle activity fuled by glucose, as we have reported previously [26]. Time-dependent depletion of the 12C (unlabeled) counterparts (● versus ●) was also evident, which indicates the disruption of the Krebs cycle fueling by other substrates (unlabeled in this case) such as Gln described above. We further noted depletion of 13C3- and/or 13C1-isotopologues of citrate (B), malate (D), and Glu (F) by selenite, which can be accounted for by blocked pyruvate carboxylase (● PCB) and malic enzyme (● ME) activities, respectively [13, 15].
We thus examined time course PCB protein expression in response to selenite and found that inhibition of PCB activity was elicited by reduced protein expression (I-J). In addition to perturbed Krebs cycle functions, selenite induced accumulation of 13C3-Ser (G) but depletion of 13C3-Ala (H), which could reflect enhanced Ser but blocked Ala biosynthesis. The latter should contribute to decreased 13C-Ala incorporation into proteins (K). Reduced de novo protein synthesis may also be occurring, as evidenced by the accumulation of all essential amino acids such as phenylalanine (Phe) and valine (Val) (L and data not shown). Although less in extent, 13C1-Ser accumulation were also noted. This 13C scrambled species could reflect impact of selenite on the pentose phosphate pathway (PPP) and/or gluconeogenesis (GNG) [27].
To verify GNG and ME activity in A549 cells, we examined 13C incorporation into Ser in the labeled Gln time course experiment described above. As Figure S2 shows, 13C was incorporated into Ser from [13C5,15N2]-Gln, which requires at least part of the GNG to produce 3-phosphoglycerate (3-PGA) as precursor to Ser synthesis. The m+1 and m+2 isotopologues in GC-MS data were assigned to 15N1-Ser and [13C1,15N1]-Ser, respectively based on transamination-requiring Ser synthesis from [13C6]-glucose (Fig. S1H). As the case for the glucose experiment, we observed enhanced Ser synthesis from Gln induced by selenite. We also saw similar time-dependent decrease in the levels of 13C5,15N1-/13C5-Glu (A) and 13C4-malate (C) in response to selenite as in Fig. S1A,C, which indicates inhibition of the GLS and PDH-initated Krebs cycle activity (●). The presence of 13C2- and 13C3-Glu (B) -malate (D), -citrate (F) indicates the activity of ME-PDH (●,●) and ME-PCB (●) pathways (Fig. S2). Again, selenite treatment led to dramatic depletion of these labeled species, which agrees with the blocked ME, PCB, and PDH activity, as evident in Fig. S1. Thus, selenite can inhibit A549 cell growth by disrupting multiple input reactions for the Krebs cycle, including GLS, PDH, PCB, and ME activities.
B. Ex vivo human lung tissue models
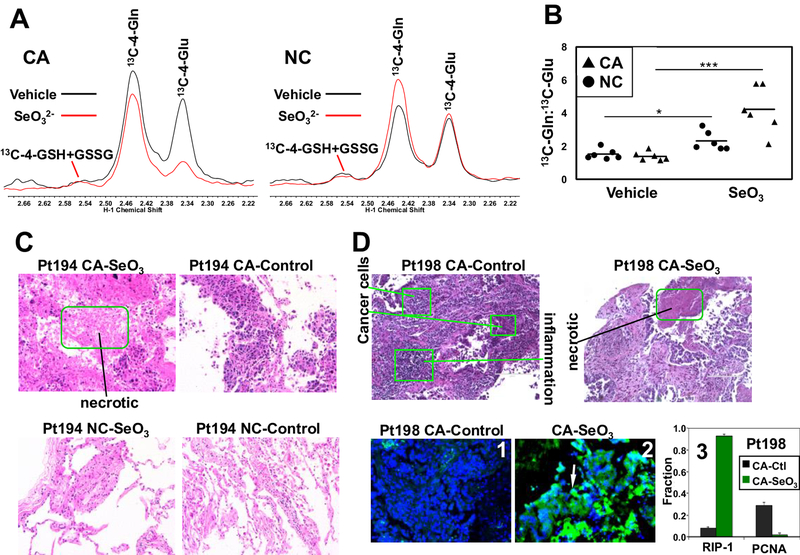
We next tested whether selenite inhibits glutaminolysis in paired CA and NC lung tissue slice cultures freshly prepared from patients with primary NSCLC. This is a much more clinically relevant model than 2D cell cultures and enables comparison of CA versus NC tissues on an individual patient basis [19, 28]. Tissue slice cultures were treated with or without 6.25 μM Na2SeO3 in the presence of [13C5,15N2]-Gln for 24 h. Consistent with the cell experiments, selenite induced a substantial depletion of 13C-Glu in the CA tissues but much less so in the NC tissue slices (Fig. 2A). Quantification of 13C-Gln and -Glu yielded signicantly elevated Gln/Glu ratios (about 2-fold higher) in six pairs of selenite-treated CA versus NC tissues, consistent with greater impairment of glutaminolysis in CA than NC tissues (Fig. 2B). We further examined the histopathology of two pair of treated tissue slices using hematoxylin-eosin (H&E) and immunofluorescent (IF) staining. As shown in Fig. 2C, the CA tissue slice of NSCLC patient 194 responded to selenite with extensive tissue necrosis while the NC counterpart showed little change. This necrotic response also occurred in selenite-treated CA tissue slice of another patient (198) by both H&E and IF staining for a necroptosis marker RIP-1 [29] (Fig. 2D). In addition to dramatic increase in the RIP-1 fluorescence (2 versus 1; 3), the PCNA (mitotic index marker) fluorescence showed opposite response to selenite (3). These results indicate that inhibition of glutaminolysis by selenite was associated with CA tissue damages.
Figure 2. Selenite impairs glutaminolysis in NSCLC tumor tissues.
A) 1D 1H{13C} HSQC spectra were recorded from polar extracts of a matched pair of patient-derived NC and CA lung tissue slices treated with [13C5,15N2]-Gln tracer ± 6.25 μM Na2SeO3 for 24 h. Control and selenite spectra were superimposed and normalized to total protein to visualize differences between treatments in the areas of the 13C-4-Gln and 13C-4-Glu peaks at 2.34 and 2.44 ppm, respectively. B) peak area ratios 13C-4-Gln:13C-4-Glu were calculated from HSQC spectra as shown in A for a total of 6 patients. ***p < 0.001, *p < 0.05 for comparing the 13C-Gln:13C-Glu ratio of selenite versus vehicle treatments in CA and matched NC tissue slices, respectively. The p value for the comparison of the selenite-treated cancer versus non-cancer pairs was 0.0169. C) H&E staining of an example pair of CA and matched NC tissues after the tracer and selenite treatment in A. Green box denotes the necrotic region in selenite-treated CA tissues; images were 200x magnified. D) H&E staining of another patient’s CA tissues showing necrosis in response to selenite (top panels). Also shown is the selenite-induced increase in RIP-1 green fluorescence (2 versus 1) and the fraction of RIP-1 expressing cells (q=0.0008) as well as selenite-induced decrease in that of PCNA-expression cells (3) *** q<0.001, * q<0.05.
GLS1 inhibitor CB-839 elicits similar metabolic responses as selenite
To determine whether inhibition of glutaminolysis can lead to similar growth and/or metabolic responses as the selenite treatment in the two lung cancer models, we performed experiments with CB-839 treatment using 13C5,15N2-Gln as tracer on A549 cells for 24–72 h and CA lung tissue slices for 24 h. CB-839 is a highly specific and potent inhibitor of GLS1 [30]. As shown in Figure S3A, 24 h of CB-839 treatment induced a buildup of 13C-Gln and depletion of the 13C-Glu product as well as its subsequent transformations into glutathiones (13C-GSH+GSSG), the Krebs cycle metabolites (13C-citrate, -Asp), and nucleotides (13C-UTP) in A549 cells, which were also noted in 48–72 h of treatments. These metabolic effects were accompanied by growth inhibition (inset) and were similar to those of the selenite treatment (cf. Fig. 1). Moreover, 13C-Gln buildup and 13C-Glu depletion were evident in an NSCLC patient’s CA tissue slices (Fig. S3B), showing similar qualitative responses to CB-839 between A549 cells and lung cancer tissue. The CB-839 metabolic data are also qualitatively similar to the effects of selenite in tissue slices (see below). These data are consistent with inhibition of GLS1 activity in both lung cancer models in response to selenite.
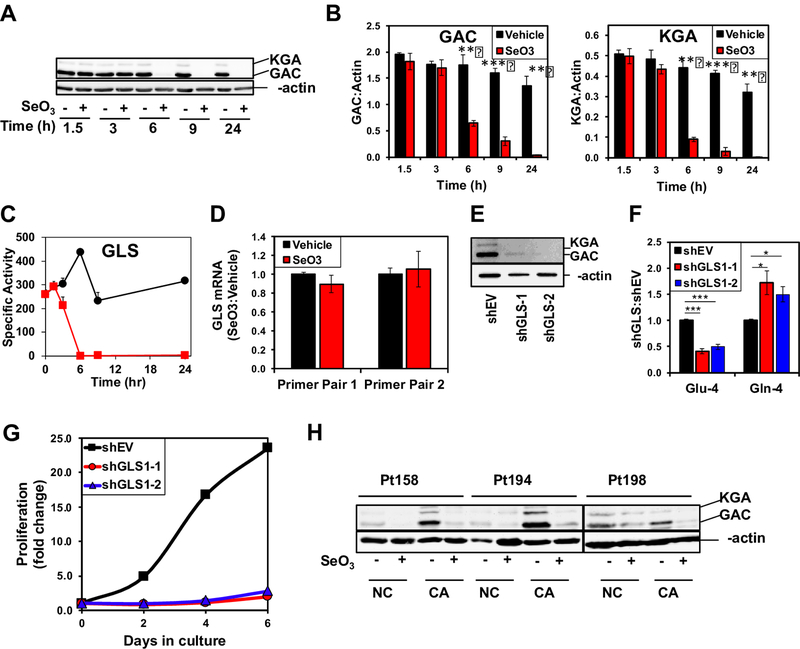
Selenite inhibits GLS activity by suppressing GLS1 protein expression
We then asked if selenite inhibits GLS activity by suppressing the protein expression. The two splice variants, kidney-type (KGA) and glutaminase C (GAC), of GLS1 [31] are the main glutaminase isoforms in NSCLC while the liver isoform GLS2 is a minor component (data not shown). The time course for the protein expression of the two GLS1 variants was probed using an antibody that recognizes a common epitope. Both splice variant levels started to decline by 6 h and were undetectable by 24 h (A-B) (Fig. 3) so did GLS activity (C), which is consistent with the time course of metabolite measurements in Fig 1. To determine whether the decrease in GLS1 protein expression resulted from gene suppression by selenite, we measured GLS1 transcript levels by qRT-PCR. No significant changes in GLS1 transcript levels in A549 cells were evident using two distinct PCR primer pairs after 24 h treatment with selenite (D), suggesting that selenite decreased GLS1 expression at the protein level independent of transcriptional regulation.
Figure 3. Selenite attenuates GLS expression in A549 cells and human NSCLC tissues.
A) Time-dependent suppression of GLS1 proteins by selenite in A549 cells. A549 cells were treated with or without 6.25 μM Na2SeO3 and harvested at 0, 1.5, 3, 6, 11, and 24 h after treatment for Western blot analysis. Shown is a representative blot. B) Quantification of GAC and KGA intensities in Western blots normalized to that of β-actin p<0.001 *** p<0.01 ** . C) Time course inhibition of GLS activity by selenite in A549 cells. D) Lack of selenite effect on GLS1 gene expression. A549 cells were treated with or without 6.25 μM Na2SeO3 and harvested at 24 h after treatment. Total RNA was extracted and GLS1 gene expression measured using qRT-PCR. ΔΔCt values were calculated with 18S rRNA as housekeeping gene and the ratios of these values for selenite versus control samples were shown for two primer sets. E) Silencing of GLS1 protein expression in A549 cells using two separate shRNA vectors (shGLS’s). Western blot analysis of lysates showed knockdown efficiency for GLS1 shRNA’s against the empty vector (shEV). F) Inhibition of 13C-4-Gln converstion to 13C-4-Glu in 13C5,15N2-Gln-treated GLS1 knockdown cells for 24 h. 13C labeled Gln and Glu were quantified using 1H{13C} HSQC NMR as in Fig. 2. *q<0.05; ***q<0.001; n=3. G) Growth inhibition by GLS1 suppression in A549 cells. H) Suppression of GLS1 proteins by selenite in CA and adjacent NC lung tissue slices. Three matched pairs of CA and NC were treated with or without 6.25 μM Na2SeO3 for 24 h, extracted for proteins, and analyzed by Western blotting, as described in methods.
To confirm that GLS1 mediated hydrolysis of Gln to Glu in A549 cells, we used two shRNAs to suppress GLS1 expression, which resulted in near complete suppression of KGA and GAC protein variants with both shRNA constructs (E) (Fig. 3). We then measured glutaminolysis in A549 cells by monitoring the uptake and conversion of [13C515N2]-Gln to the [13C]-Glu products. Suppression of GLS1 by shRNAs led to a qualitatively similar effect as selenite treatment with a buildup of [13C]-Gln and depletion of [13C]-Glu, (F). Additionally, A549 cell proliferation was dramatically inhibited by prolonged GLS1 knockdown (G), which is akin to the growth effect of prolonged selenite treatment (cf. Fig. 1 in [11]). These experiments confirm that GLS1 catalyzes a significant fraction of glutamate production in A549 cells and supports the finding that selenite inhibits glutaminolysis through GLS1 depletion.
We next measured GLS1 expression in response to selenite in human NSCLC tissues. As in A549 cells, selenite decreased GLS1 protein levels in both CA and NC tissues from three responsive patients (Fig. 3H). This should lead to a comparable, selenite-induced increase in the 13C-Gln/Glu ratio for both tissue types. The higher 13C-Gln/Glu ratio observed in CA tissues (Fig. 2B) suggests that CA tissues rely more heavily on GLS activity for glutamate production and subsequent anaplerotic input into the Krebs cycle than do NC tissues.
Selenite increases the oxidized:reduced glutathione ratio
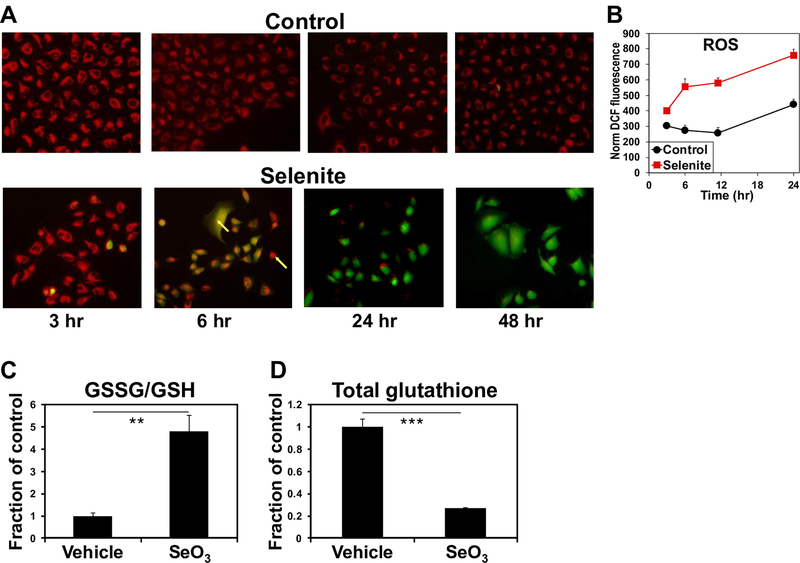
As selenite is known to induce oxidative stress, we assessed ROS status by measuring the DCFDA fluorescence following selenite treatment in A549 cells. DCFDA fluorescence began to increase at 3 h and continued to rise over the course of 48 h (Fig. 4A,B). We then used 1H NMR to measure the ratio of oxidized to reduced glutathione (GSSH:GSH) in control and selenite-treated A549 cell extracts as another measure of oxidative stress. The GSSG:GSH ratio increased by ~4 fold in response to 24 h of selenite treatment (Fig. 4C), consistent with the induction of oxidative stress by selenite. We also note from the NMR analysis that selenite greatly reduced the total (Fig. 4D) and de novo synthesized glutathione pools (Fig. 1D). These agreed with the suppression of GLS1 expression, as its product glutamate is a precursor for glutathione synthesis. Taken together these results suggest that selenite induces oxidative stress at least in part via decreasing the capacity of A549 cells to detoxify reactive oxygen species (ROS) by inhibiting glutamate-dependent glutathione synthesis.
Figure 4. Selenite induces oxidative stress in A549 cells.
A-B) Time course ROS production induced by selenite. A549 cells were treated with or without 6.25 μM Na2SeO3 and prepared for ROS measurement at 3, 6, 24, and 48 h after treatment as described in Method. In A, red fluorescence indicated mitochondria and green fluorescence reported DCF as a measure of ROS production. Co-localization of the mitochondrial and ROS markers was evident at the 6 h time point. In B, the DCF fluorescence was quantified by microplate reader in parallel. C-D) Selenite increased GSSG/GSH ratios and attenuated total glutathione pool. A separate experiment was performed for acquiring 1D 1H NMR spectra from the polar extracts of A549 cells treated with or without 6.25 μM Na2SeO3 for 24 h. GSSG and GSH were quantified from their cysteine-2 residue at 4.75 and 4.56 ppm, respectively. Shown in C are the GSSG:GSH ratios for selenite versus vehicle. **p<0.01; n=3. Total glutathione was quantified from the glutamate-4 residue of GSSG+GSH at 2.55 ppm in D. ***p<0.001; n=3.
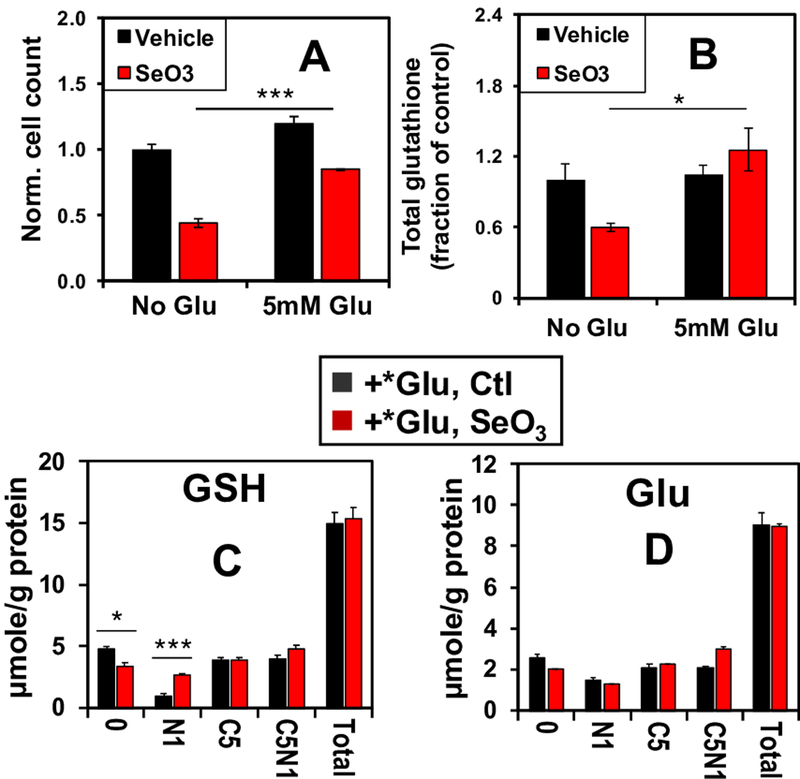
Glutamate supplementation rescues selenite-induced growth inhibition and glutathione depletion
To further test the hypothesis that decreased GLS1 expression and consequent depletion of Glu induced by selenite was responsible for the decreased total glutathione levels, increased GSSG/GSH ratios, and decreased proliferation, we treated A549 cells with 6.25 μM Na2SeO3 in the presence or absence of 5 mM Glu. By supplying the cells with the direct product of the GLS1 reaction, we expected to mitigate the anti-proliferative effects of selenite. Indeed, Glu supplementation rescued a large fraction of the selenite-induced inhibition of A549 cell proliferation (Fig. 5A). It also rescued selenite-induced depletion of total glutathione (Fig. 5B). Moreover, to confirm that selenite directly enhanced the incorporation of extracellular Glu into glutathione, we treated cells with 5 mM [13C5,15N1]-Glu ± selenite for 24 h, and measured the enrichment of 13C and 15N labeled glutathione isotopologues using IC-UHR FT-MS. Incorporation of the extracellular [13C5,15N1]-Glu tracer into glutathione is expected to produce the [13C5,15N1]-glutathione isotoplogue. As seen in Figure 5C, Glu supplementation in selenite-treated cells restored the levels of this (C5N1) and [13C5] (C5) isotopologues as well as the total level (total) to those in the vehicle control. Similar restoration was observed in the levels of the corresponding isotopologues of the precursor Glu (Fig. 5D). The levels of 13C- and/or 15N-isotopologues of ATP and UTP as well as the total levels were also fully or largely restored, which parallels the growth rescue. However, exogenous Glu significantly enhanced the levels of [15N1]-GSH (Fig. 5C), [13C5]-αKG (Fig. S4A) and [13C4]-succinate (Fig. S4B) while failing to restore those of [13C4]-fumarate (S4C), -malate (S4D), -Asp (S4E) and -citrate (S4F); neither did it restore the total levels of the latter metabolites in response to selenite treatment. The changes in the Krebs cycle metabolites are consistent with additional block(s) in the Krebs cycle, possibly at the SDH step that can not be overcome by Glu supplementation. It is plausible that selenite also enhanced the transamination of αKG leading to [15N1]-GSH accumulation. Altogether, these results suggest that selenite toxicity in A549 cells involves multiple targets but arises largely from glutamate depletion via GLS1 suppression and reduced glutaminolysis.
Figure 5. Glutamate supplementation reverses selenite-induced growth inhibition and glutathione depletion in A549 cells.
A549 cells were grown with or without 6.25 μM selenite in combination with 5 mM Glu supplement for 24 h before fixation, DAPI staining, and automated cell counting. Shown in A are the cell count ratios of treatments versus PBS (no-Glu control). ***p<0.001. A parallel experiment was performed for the extraction of polar metabolites and 1D 1H NMR analysis. Total glutathione was quantified as in Figure 4D, and its ratios of selenite over control are shown in B (*p<0.05). C-D. A third experiment was conducted in the presence of 5 mM 13C5,15N1-Glu (*Glu) for 24 h. Polar extracts were analyzed by IC-UHR FTMS to determine the distribution of 13C and 15N labels in GSH and Glu. 0, N1, C5, C5N1, and total refer to GSH or Glu with no 13C/15N labels, 15N1, 13C5, 13C5,15N1, and sum of all isotopologues, respectively. *q < 0.05; **q <0.01
Discussion
Selenite has been shown recently by Zhao et al. [8] to suppress GLS1 levels via enhancing the ubiquitin-dependent degradation pathway in colorectal cancer cells. However in this study, no direct evidence for reduced GLS activity was shown, nor was metabolic mechanism examined to account for seleinite-induced growth inhibition. As enzymic acivities in situ are governed by multiple factors including protein levels, post-translational modifications, allosteric modulation, and substrate availability via compartmentation [32], reduced protein expression alone does not necessarily predict metabolic outcomes.
Here, we employed SIRM to provide direct evidence for selenite inhibition of GLS activity, which was linked to GLS suppression in human lung cancer cells and ex vivo patient tumor tissues. Our SIRM studies also revealed metabolic mechanisms underlying selenite-induced growth inhibition and excess ROS production. This was shown first by observing large Gln accumulation and Glu depletion (Fig. 1), which is consistent with decreased GLS activity. Reduced GLS activity was then confirmed by tracing the conversion of [13C5,15N2]-Gln to 13C labeled Glu products and GLS activity assays. Such reduced glutaminolysis in turn blocked anaplerotic input into the Krebs cycle and its activity (Fig. S2). The depletion of labeled glutaminolytic products (glutathione in particular) and Krebs cycle activity began at 3–6 h of selenite treatment, which is commensurate with the occurrence of ROS (Fig. 4A–B). This supported the notion that excess ROS accumulation results from the dysfunctioning of the Krebs cycle (Fig. S2) and glutathione depletion (Fig. 4). Selenite-induced inhibition of glutaminolysis in A549 cells resulted from the suppression of GLS1 protein (instead of gene) expression, as the protein level decreased precipitously after 6 h of treatment while the transcript level was unaltered (Figs. 1 and 3B,D).
More importantly, the in vitro cell-based findings recapitulated those of the more clinically relevant ex vivo human tissue culture studies, where selenite inhibited glutaminolysis and GLS1 protein expression, while inducing necroptosis in patients’ CA lung tissues (Fig. 2). It is also interesting to note that although GLS1 expression was also reduced in the NC counterparts (Fig. 3H), neither Gln to Glu conversion nor histopathology was significantly affected. This implies that NC tissues are not “addicted” to GLS1-cataylzed glutaminolysis. We surmise that less proliferative NC tissues do not require high anaplerotic activities and are thus less dependent on GLS1 for Glu production. Other pathways such as amidotransferase reactions and the urea cycle could contribute more to Gln to Glu conversion under both control and selenite treatments. Further studies will be needed to verify this hypothesis.
Further confirmation for the key role of GLS1 in mediating selenite toxicity in A549 cells came from the GLS1 KD and Glu rescue experiments. GLS1 KD suppressed glutaminolysis and proliferation (Fig. 3E–G) while Glu supplementation rescued the two processes (Fig. 5). It should be noted that Glu-mediated rescue did not fully restore the Krebs cycle activity (Fig. S4) and growth. This may well be caused by ability of selenite to block the PCB and ME activities (Figs. S1–2), which we have shown previously to be important for A549 cell growth both in vitro and in vivo [12, 13],[33]. Interestingly, enhanced PCB expression in pancreatic cancer cells is associated with adaptive resistance to CB-839 inhibition of glutaminolysis [34].
In conclusion, we have demonstrated the metabolic mechanism of selenite toxicity in both lung cancer cells in vitro and patient lung cancer tissues ex vivo. The toxicity was mediated by suppressing GLS1 protein expression together with inhibition of PCB and ME activity, followed by disruption of the Krebs cycle activity and glutathione synthesis. These effects of selenite on multiple anaplerotic targets could reduce the likelihood of resistance development in lung cancer cells.
Supplementary Material
Statement of significance.
Selenium-containing compounds have long been investigated for their chemopreventive and therapeutic properties in cancer, often with conflicting results. These conflicts may result from using chemical forms of selenium, as they have widely different mechanisms of toxicity and bioavailability. Here, we investigate inorganic selenite, a common dietary supplement and naturally occurring in foods, as a potential therapeutic agent for non-small cell lung cancer (NSCLC). We demonstrate that selenite inhibits glutaminolysis by decreasing glutaminase expression, leading to increased oxidative stress by depleting glutamate-dependent glutathione synthesis in lung cancer cells and tissues. These results represent a novel mechanism for selenite-induced toxicity and oxidative stress and may be exploited clinically for treatment of glutamine-dependent NSCLCs.
Acknowledgements
We thank Dr. Michael Bousamra for performing the surgery and tissue resection, JinLian Tan for her assistance in the patient tissue slices experiments, and Dr. Maria Scavo for performing the IF staining of tissue slices. We thank Dr. Susan Demo of Calithera for the generous gift of CB-839.
Financial Support: This work was supported in part by grants: NIH 1R01CA118434–01A2 (TWMF), 3R01CA118434–02S1 (TWMF), 1P01CA163223–01A1 (to ANL and TWMF), 1U24DK097215–01A1 (to RMH, TWMF, and ANL), and P30CA177558 (to B.M. Evers). RCB was supported by a T32 CA165990 (V. Ragnekar).
ABBREVIATIONS
- CA
Cancer
- DTT
dithiothreitol
- H2-DCFDA
2’,7’-dichlorodihydrofluorescein diacetate
- DMEM
DSS 4,4 dimethylsilapentane-1-sulfonate; Dulbecco’s Modified Eagle’s Medium
- FDR
false discovery rate
- FBS
fetal bovine serum
- FWHM
full width at half maximum
- GC-MS
gas chromatography-mass spectrometry
- GNG
gluconeogenesis
- GLS
Glutaminase
- KGA
kidney-type glutaminase
- GAC
glutaminase C
- GSSG
oxidized glutathione
- GSH
reduced glutathione
- H&E
hematoxylin and eosin
- HSQC
heteronuclear single quantum coherence
- IF
immunofluorescence
- IC-UHR FT-MS
Ion chromatography-ultra high-resolution Fourier transform-MS
- ME
malic enzyme
- NAC
N-acetylcysteine
- NC
Non-cancerous
- NSCLC
Non-Small Cell Lung Cancer
- MTBSTFA
N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide
- PD
Patient-derived
- PCB
pyruvate carboxylase
- PDH
pyruvate dehydrogenase
- PPP
Pentose Phosphate Pathway
- PBST
phosphate buffered saline tween20
- ROS
Reactive Oxygen Species
- SDS
sodium dodecyl sulfate
- SIRM
Stable-Isotope Resolved Metabolomics
- TBDMSCl
tert-Butyldimethylchlorosilane
Footnotes
Conflict of Interest Statement
The authors have declared no conflicts of interest.
References
- [1].Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR, EMBO J 1986, 5, 1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Holben DH, Smith AM, J Am Diet Assoc 1999, 99, 836. [DOI] [PubMed] [Google Scholar]
- [3].Vanderpas JB, Contempre B, Duale NL, Goossens W, Bebe N, Thorpe R, Ntambue K, Dumont J, Thilly CH, Diplock AT, Am J Clin Nutr 1990, 52, 1087; Y. Y. Cheng, P. C. Qian, Biomed Environ Sci 1990, 3, 422; E. W. Taylor, Biol Trace Elem Res 1995, 49, 85. [DOI] [PubMed] [Google Scholar]
- [4].Dunn BK, Richmond ES, Minasian LM, Ryan AM, Ford LG, Nutr Cancer 2010, 62, 896. [DOI] [PubMed] [Google Scholar]
- [5].Brozmanova J, Manikova D, Vlckova V, Chovanec M, Arch Toxicol 2010, 84, 919. [DOI] [PubMed] [Google Scholar]
- [6].Clark LC, Dalkin B, Krongrad A, Combs GF Jr., Turnbull BW, Slate EH, Witherington R, Herlong JH, Janosko E, Carpenter D, Borosso C, Falk S, Rounder J, Br J Urol 1998, 81, 730. [DOI] [PubMed] [Google Scholar]
- [7].Yamaguchi K, Uzzo RG, Pimkina J, Makhov P, Golovine K, Crispen P, Kolenko VM, Oncogene 2005, 24, 5868; S. Reagan-Shaw, M. Nihal, H. Ahsan, H. Mukhtar, N. Ahmad, Prostate 2008, 68, 1624; S. D. Cho, C. Jiang, B. Malewicz, Y. Dong, C. Y. Young, K. S. Kang, Y. S. Lee, C. Ip, J. Lu, Mol Cancer Ther 2004, 3, 605; D. P. Cherukuri, M. A. Nelson, Cancer Biol Ther 2008, 7, 697. [DOI] [PubMed] [Google Scholar]
- [8].Zhao J, Zhou R, Hui K, Yang Y, Zhang Q-Y, Ci Y, Shi L, Xu C, Huang F, Hu Y, Oncotarget 2017, 8, 18832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Park SH, Kim JH, Chi GY, Kim GY, Chang YC, Moon SK, Nam SW, Kim WJ, Yoo YH, Choi YH, Toxicol Lett 2012, 212, 252. [DOI] [PubMed] [Google Scholar]
- [10].Ramoutar RR, Brumaghim JL, Cell Biochem Biophys 2010, 58, 1. [DOI] [PubMed] [Google Scholar]
- [11].Fan T, Bandura L, Higashi R, Lane A, Metabolomics 2005, 1, 325. [Google Scholar]
- [12].Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM, Molecular cancer 2009, 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sellers K, Fox MP, Bousamra M 2nd, Slone SP, Higashi RM, Miller DM, Wang Y, Yan J, Yuneva MO, Deshpande R, Lane AN, Fan TW, J Clin Invest 2015, 125, 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P, Signoretti S, Billiard J, Duffy KJ, Grant A, Wang X, Lorkiewicz PK, Schatzman S, Bousamra M 2nd, Lane AN, Higashi RM, Fan TW, Pandolfi PP, Sukhatme VP, Seth P, Cell Metab 2014, 19, 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fan TW, Warmoes MO, Sun Q, Song H, Turchan-Cholewo J, Martin JT, Mahan A, Higashi RM, Lane AN, Cold Spring Harb Mol Case Stud 2016, 2, a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, Chen X, Bishop JM, Cell Metab 2012, 15, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Broers JL, Viallet J, Jensen SM, Pass H, Travis WD, Minna JD, Linnoila RI, American journal of respiratory cell and molecular biology 1993, 9, 33; M. P. Fox, A. N. Lane, M. Bousamra, T. Fan, Journal of the American College of Surgeons 2011, 213, S39. [DOI] [PubMed] [Google Scholar]
- [18].Fan TW, Higashi RM, Lane AN, Drug Metab Rev 2006, 38, 707. [DOI] [PubMed] [Google Scholar]
- [19].Fan TW, Lane AN, Higashi RM, Bio Protoc 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, Zimmerman LJ, Liebler DC, Slebos RJ, Lorkiewicz PK, Higashi RM, Fan TW, Dang CV, Cell Metab 2012, 15, 110; T. W.-M. Fan, in The Handbook of Metabolomics: Pathway and Flux Analysis, Methods in Pharmacology and Toxicology., (Eds: T. W.-M. Fan, A. N. Lane, R. M. Higashi), Humana Press, New York City, U.S.A. 2012, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yang Y, Fan WW-M, Lane AN, Higashi RM, Anal. Chim. Acta 2017, 976, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fan TW-M, Lane AN, Progress in Nuclear Magnetic Resonance Spectroscopy 2008, 52; T. W.-M. Fan, in Methodologies for Metabolomics: Experimental Strategies and Techniques, (Eds: N. W. Lutz, J. V. Sweedler, R. A. Wevers), Cambridge University Press, Cambridge, United Kingdom 2013, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Moseley HN, BMC Bioinformatics 2010, 11, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Quesada AR, Sanchez-Jimenez F, Perez-Rodriguez J, Marquez J, Medina MA, Nunez de Castro I, Biochem J 1988, 255, 1031; M. O. Yuneva, T. W.-M. Fan, T. D. Allen, R. M. Higashi, D. V. Ferraris, T. Tsukamoto, J. M. Matés, F. J. Alonso, C. Wang, Y. Seo, X. Chen, J. M. Bishop, Cell Metabolism 2012, 15, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Benjamini Y, Hochberg Y, Journal of the Royal Statistical Society. Series B (Methodological) 1995, 57, 289. [Google Scholar]
- [26].Fan TWM, Bandura LL, Higashi RM, Lane AN, Metabolomics 2005, 1, 325. [Google Scholar]
- [27].Sun RC, Fan TWM, Deng P, Higashi RM, Lane AN, Le A-T, Scott TL, Sun Q, Warmoes MO, Yang Y, Nature communications 2017, 8, 1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lane AN, Higashi RM, Fan TW, Metabolomics 2016, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK, Cell 2009, 137, 1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, Janes JR, Laidig GJ, Lewis ER, Li J, Mackinnon AL, Parlati F, Rodriguez ML, Shwonek PJ, Sjogren EB, Stanton TF, Wang T, Yang J, Zhao F, Bennett MK, Mol Cancer Ther 2014, 13, 890. [DOI] [PubMed] [Google Scholar]
- [31].Cassago A, Ferreira AP, Ferreira IM, Fornezari C, Gomes ER, Greene KS, Pereira HM, Garratt RC, Dias SM, Ambrosio AL, Proc Natl Acad Sci U S A 2012, 109, 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lane AN, Fan TW, Higashi RM, Biophysical Tools for Biologists. 2008, 84, 541. [Google Scholar]
- [33].Ren JG, Seth P, Clish CB, Lorkiewicz PK, Higashi RM, Lane AN, Fan TWM, Sukhatme VP, Scientific Reports 2014, 4, 5414; K. Sellers, M. P. Fox, M. Bousamra, S. Slone, R. M. Higashi, D. M. Miller, Y. Wang, J. Yan, M. O. Yuneva, R. Deshpande, A. N. Lane, T. W.-M. Fan, J. Clin. Invest. 2015, 125, 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Biancur DE, Paulo JA, Malachowska B, Quiles Del Rey M, Sousa CM, Wang X, Sohn ASW, Chu GC, Gygi SP, Harper JW, Fendler W, Mancias JD, Kimmelman AC, Nature communications 2017, 8, 15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.