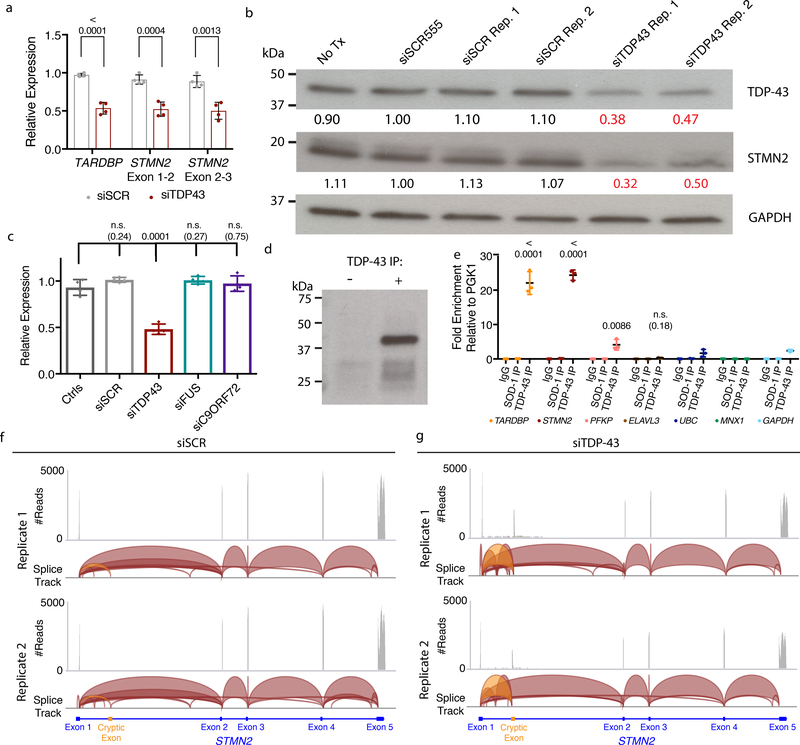
Figure 3. TDP-43 regulates STMN2 RNA and protein levels through direct interactions and suppression of a cryptic exon.
(a) qRT-PCR analysis for the STMN2 transcript using two different sets of exon spanning primer pairs. Data are displayed as mean with SD of two technical replicates from n=2 independent experiments (Unpaired t-test, two-sided, P value < 0.05). (b) Immunoblot analysis for TDP-43 and STMN2 protein levels following partial depletion of TDP-43 by siRNA knockdown. Protein levels were normalized to GAPDH and are expressed relative to the levels in hMNs treated with the siRED control. Similar results were obtained in n=2 independent experiments. (c) qRT-PCR analysis for STMN2 transcript analysis in Hb9::GFP+ hMNs treated with siRNAs targeting three ALS-linked genes (TARDBP, FUS, and C9ORF72). Data are displayed as mean with SD of replicates from n=2 independent experiments (Dunnett’s multiple comparison test, Alpha value < 0.05). Similar results were obtained in n=3 independent experiments. (d-e) Formaldehyde RNA immunoprecipitation was used to identify transcripts bound to TDP-43. After TDP-43 immunoprecipitation (d), qRT-PCR analysis was used to test for enrichment of TARDBP transcripts, STMN2, PFKP, and ELAVL3 (e) normalized to the sample input then relative to IgG and the house keeping pulled down by TDP-43. Data are displayed as mean with SD of replicates for one of n=2 independent experiments (Unpaired t-test, two-sided, P value < 0.05 compared to the house keeping genes MNX1, GAPDH, and UBC). (f-g) Visualization of RNA-seq reads mapping to STMN2 from neurons treated with scrambled siRNAs (f) or siTDP43 (g). Read coverage and splice junctions are shown for alignment of the samples to the human hg19 genome. Splice ribbons from exon 1 to the cryptic exon are highlighted in orange.