Figure 4.
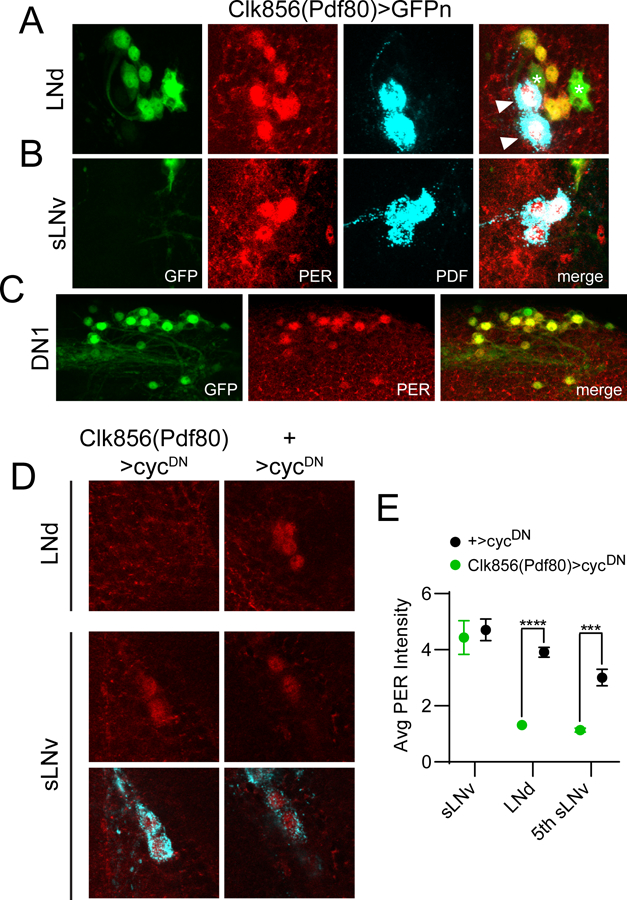
The Clk856(Pdf80) line targets non-sLNv clock cells. For panels A-C, dissections were carried out at ~ZT23 and representative confocal images are shown. GAL4 expression is visualized via a cross to UAS-nGFP (green); PER staining (red) identifies clock cells; PDF staining (cyan) labels LNv cells and processes. A, GFP staining of Clk856(Pdf80)>GFPn brains demonstrating expression in all six LNd cells. In contrast, this line lacks GAL4 expression in PDF+ sLNvs (B) but is present in the vast majority of other non-LNv clock neurons such as DN1 and DN2 cells (C). For A, note that all six PER+ LNds are co-labeled with GFP; however two PDF+ lLNvs (arrowheads) are GFP-. Note also additional expression of this GAL4 line in two PER- non-clock cells (asterisks) near the LNds. D, Representative confocal images are shown of LNd and LNv cells of Clk856(Pdf80)>cycDN flies (left) and +>cycDN control flies (right) stained for PER (red) at ZT23. PDF staining (cyan) was used to identify sLNv cells, and a merged image of PER and PDF staining is shown below each PER staining panel for the sLNvs. E, Quantification of average PER staining intensity at ZT23, normalized to background levels (mean ± SEM), for sLNv cells, LNd cells and the 5th sLNv. n= 8 brains per time point; ***p<0.001; ****p<0.0001, Sidak’s multiple comparisons test following Two-Way ANOVA.
