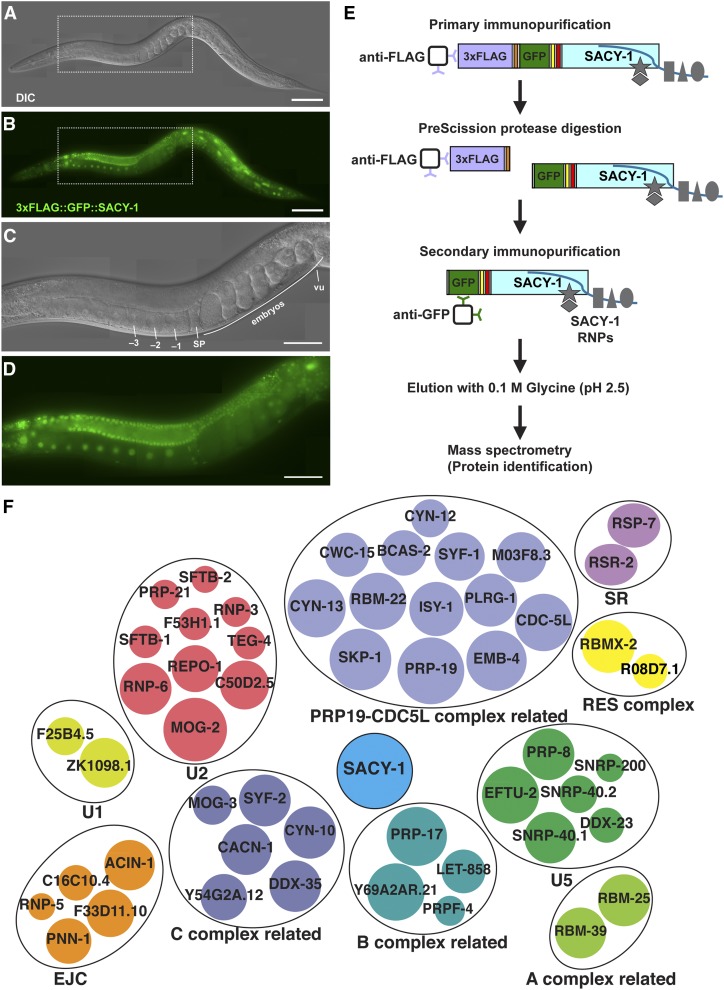
Figure 1.
Tandem affinity purification of SACY-1 to identify associated proteins. DIC (A and C) and fluorescence (B and D) photomicrographs of an adult hermaphrodite generated by CRISPR-Cas9 genome editing to express an epitope-tagged version of SACY-1 suitable for tandem affinity purification (strain DG4070 sacy-1(tn1632[3xflag::PreScission protease site::gfp::tev::s-tag::sacy-1]) I). The regions within the dotted rectangles are magnified in (C and D). Oocytes (−1, −2, −3), the spermatheca (sp), and the vulva (vu) are indicated. Bars, 100 μm in (A and B); 50 μm in (C and D). (E) Strategy for tandem affinity purification. Cleavage with PreScission protease releases GFP::SACY-1 and associated proteins from the affinity matrix, whereas the fragment with the 3xFLAG epitope and nonspecifically bound proteins are retained on the matrix. The second purification step utilized an anti-GFP affinity resin and a low pH elution. Western blot analysis of fractions from the tandem affinity purification procedure analyzed with anti-GFP or R217 anti-SACY-1(411–578) antibodies are shown in Figure S1. (F) Spliceosomal proteins reproducibly associated with SACY-1 by tandem affinity purification. Spliceosomal proteins are organized into spliceosomal subcomplexes according to Bessonov et al. (2008) or the supplemental references in Table S3. The primary data on which figure is based are presented in File S1 and Table S3. The area of each circle represents the maximal percentage coverage of each protein in a single gel slice from the purification from DG4068 (see Table S3). For example, the percent coverage of SACY-1 was 78.9% and the percent coverage of MOG-2 was 56.1%. The names of the human orthologs of the C. elegans proteins are listed in Table S3.