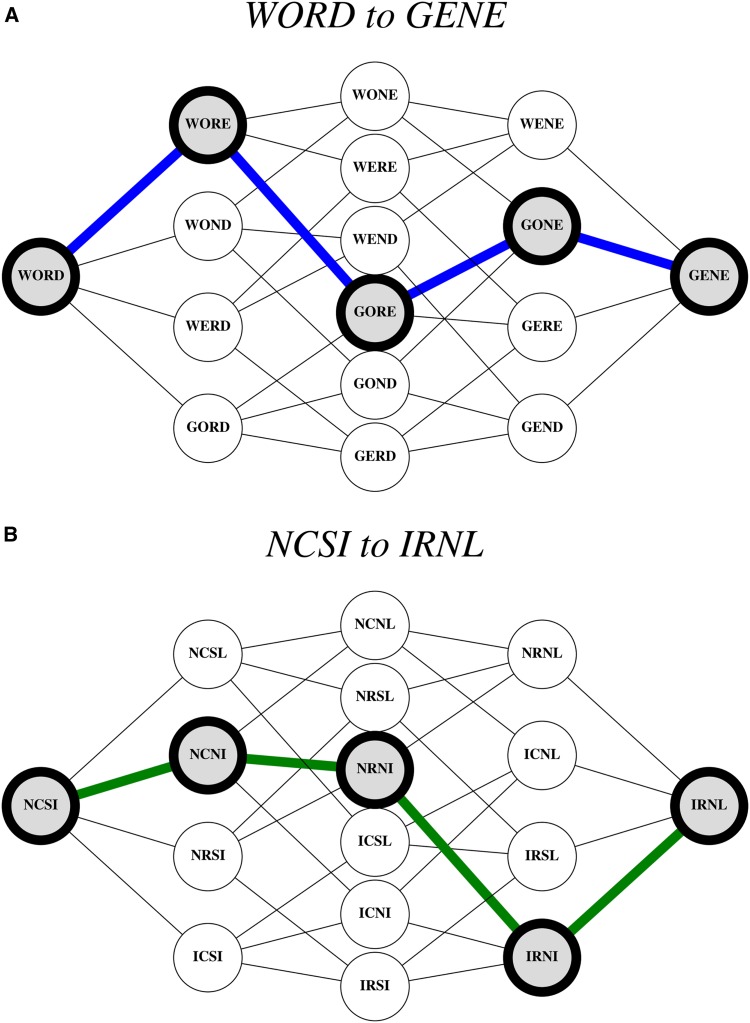
Figure 1.
Representations and applications of John Maynard Smith’s Protein Space. (A) A hypercube “fitness graph” representation, where WORD and GENE are the terminal nodes, and there exists a pathway between them (dense blue line, left to right). Importantly, all words along the pathway are sensical English words. (B) A hypercube representation of an empirical fitness landscape corresponding to mutations in dihydrofolate reductase, an enzyme target of drugs in many microbial diseases. Specifically, the mutations are associated with resistance to pyrimethamine, an antimicrobial drug. Letters correspond to single-letter amino acid abbreviations and their substitutions: asparagine (N) to isoleucine (I), cysteine (C) to arginine (R), serine (S) to asparagine (N), and isoleucine to leucine (L). The NCSI node represents the wild type and the IRNL represents the quadruple mutant with much higher resistance to the drug. The dense green line corresponds to the most likely path of enzyme evolution under selection at high concentrations of pyrimethamine: NCSI → NCNI → NRNI → IRNI → IRNL [for more details, see Ogbunugafor and Eppstein (2016)].