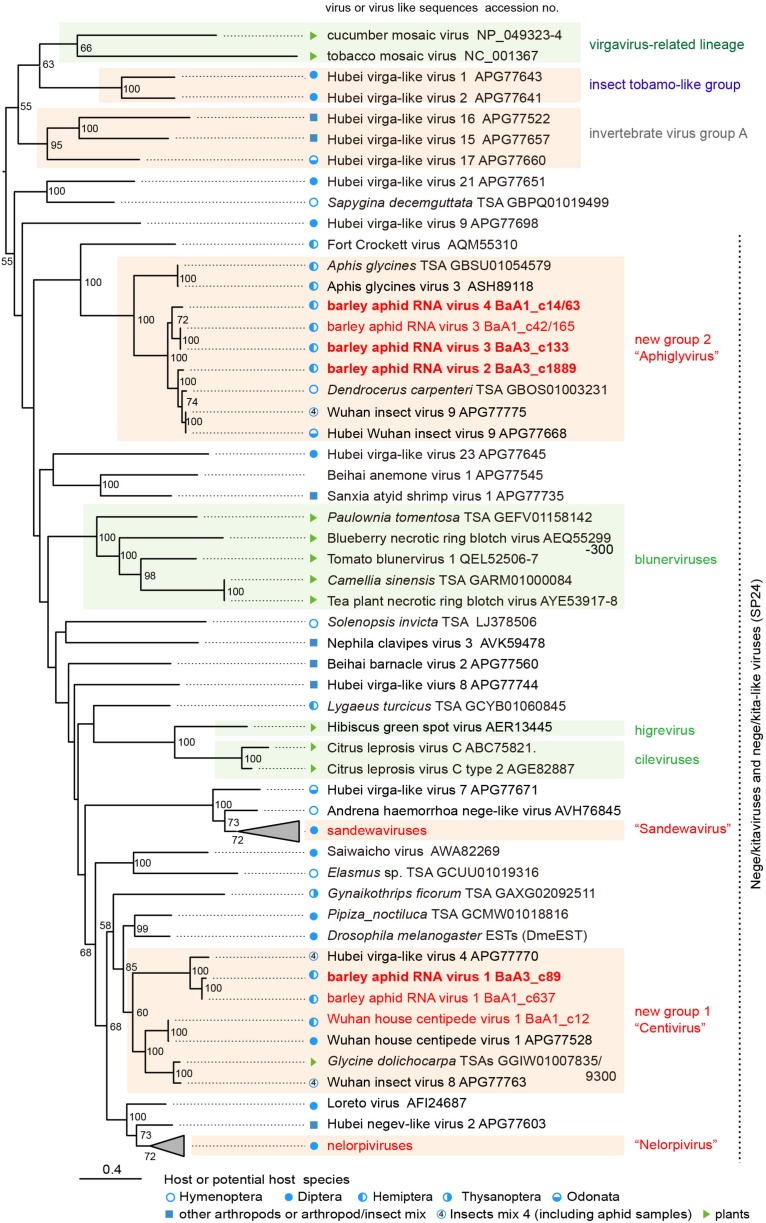
FIGURE 3.
Phylogenetic relationships of the nega/kitaviruses and their related viruses and other virus-like sequences. The maximum likelihood (ML) phylogenetic tree was constructed using PhyML 3.0 based on a multiple amino acid sequence alignment of the replicase protein or its candidate sequences. A model LG + I + G + F was selected as a best-fit model for the alignment. The tree was rooted using the midpoint rooting method. Virus names referring to plant-infecting viruses (genera Cilevirus, Higrevirus, and Blunervirus, family Kitaviridae) and insect-infecting or associated viruses are followed by the GenBank accession numbers of their sequences. The selected members of the proposed groups “Sandewavirus” and “Nelorpivirus” are displayed as collapsed triangles (see Supplementary Table S3 for the virus names in the collapsed triangles). Nege/kitavirus-like transcriptome shotgun assembly (TSA) sequences, discovered by this study (see Supplementary Table S2) or our previous study (Kondo et al., 2019), were also included in this analysis. A library of insect mixtures (WHCCII, insect virus 4) for the meta-transcriptomic analysis included two aphid species, a parasitoid wasp, and several other insects (Shi et al., 2016a). The hosts, or possible hosts, for nege/kitaviruses or virus-like sequences present in TSAs are marked with symbols (circles, squares, and triangles, respectively). Vertical dashed lines indicate that most of those viruses or virus-like sequences potentially encode conserved small proteins (SP24 family) (Kuchibhatla et al., 2014). The selected members of distantly related virus lineage, belonging to the plant virgavirus-related lineage (tobacco mosaic and cucumber mosaic viruses), insect tobamo-like virus group (Hubel virga-like viruses 1 and 2), and invertebrate virus group A (Hubel virga-like viruses 15, 16, and 17) (Kondo et al., 2019), were used as the outgroups. The scale bar represents amino acid distances. The numbers at the nodes are bootstrap values of >50%.