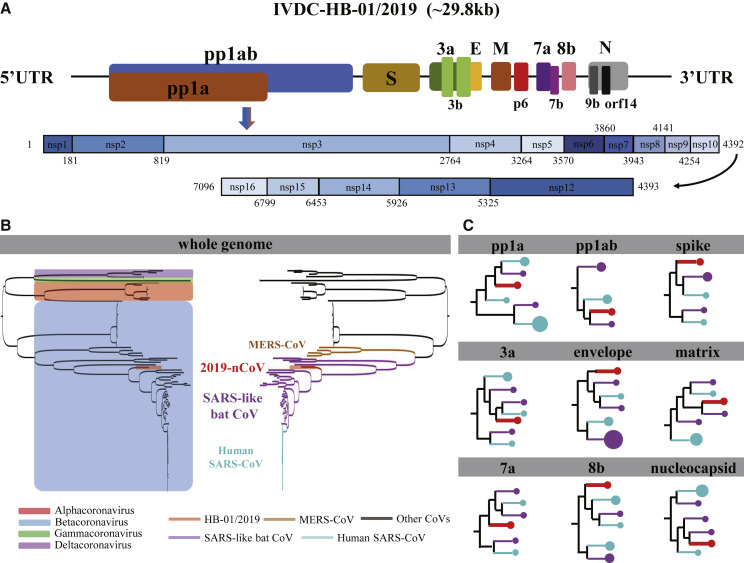
Figure 1.
Genome composition and phylogenetic tree for 2019-nCoV
(A) Schematic diagram of the genome organization and the encoded proteins of pp1ab and pp1a for the IVDC-HB-01/2019 (HB01) strain. The largest gene, namely the orf1ab, encodes the pp1ab protein that contains 15 nsps (nsp1-nsp10 and nsp12-nsp16). The pp1a protein encoded by the orf1a gene also contains 10 nsps (nsp1-nsp10). Structural proteins are encoded by the four structural genes, including spike (S), envelope (E), membrane (M), and nucleocapsid (N) genes. The accessory genes are distributed among the structural genes. The protein-encoding genes of the genome of 2019-nCoV were predicted by the online servers of GeneMarkS (http://exon.gatech.edu/GeneMark/genemarks.cgi) and ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) with manual check.
(B) Phylogenetic relationship based on the whole genome for the HB01 strain and other coronaviruses. All viral strains were classified by the genus and the type, which are presented on the left and right schematic phylogenetic trees, respectively. The four genera of the coronaviruses, including Alphacoronavirus (red), Betacoronavirus (blue), Gammacoronavirus (green), and Deltacoronavirus (violet) are blocked in the left phylogenetic tree. The MERS coronavirus (brown), the SARS-like bat coronavirus (violet), human SARS coronavirus (light blue), and the HB01 strain (red) are highlighted by lines of different colors in the right phylogenetic tree.
(C) Schematic phylogenetic trees of individual genes for the HB01 strain. The coronavirus species were colored in the same way as (B). The amount of the strains in the phylogenetic clade is denoted by the area of the circles.