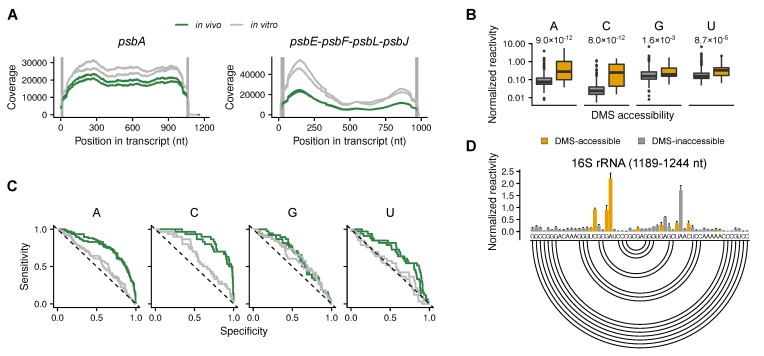
Figure 1.
Dimethyl sulfate (DMS) probing of chloroplast RNAs. (A) Examples of Illumina reads’ coverage on selected chloroplast transcripts (monocistronic psbA and polycistronic psbE-psbF-psbL-psbJ are shown) of in vivo samples (green) and in vitro-folded RNA (grey) (the same samples are presented in C). Two biological replicates each are shown. Grey, vertical lines depict the regions bound by the primers. These regions are excluded from the further analysis. For the coverage of the other transcripts see Supplemental Figure S1. (B) Normalized DMS reactivity at all four nucleotides of the 16S rRNA. The box-plots show that DMS-accessible (i.e., single-stranded and solvent-accessible) nucleotides are more likely to be modified by DMS than DMS-inaccessible (paired or solvent-inaccessible) nucleotides. Statistically significant differences were calculated using the Wilcoxon rank sum test; the resulting p-values are shown. Compare also the detected mutation rate (Supplemental Figure S2B). (C) Receiver operating characteristics (ROC) curves for the DMS reactivity profile of chloroplast 16S rRNA for all four nucleotides. (D) Normalized DMS reactivity of the in vivo samples of a selected 16S rRNA region (1189–1244 nt). Grey and yellow bars denote DMS-inaccessible and DMS-accessible nucleotides, respectively. Higher normalized DMS reactivity values indicate nucleotides that are more accessible. The secondary structure is presented by an arc-plot (bottom). Base-paired nucleotides are connected by arcs (compare results to [30]).