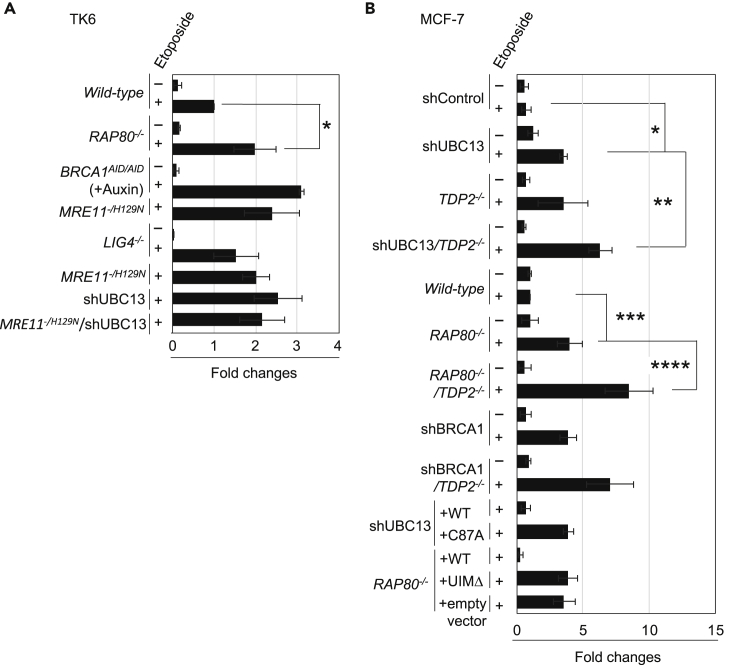
Figure 3.
UBC13, RAP80, and BRCA1 Promote the Removal of Etoposide-Induced TOP2 Adducts Independent of TDP2
(A) Quantification of TOP2-DNA-cleavage-complexes (TOP2ccs) in TK6 lymphoid cells carrying the indicated genotypes relative to the amount of TOP2ccs in wild-type cells. Schematic of in vivo TOP2cc measurement by immunodetection with α-TOP2 antibody is shown in Figure S3A. Cells were treated with etoposide (10 μM) (“+”) or DMSO (“-”) for 2 h. BRCA1AID/AID cells were pretreated with auxin for 2 h, then incubated with etoposide (10 μM) plus auxin for an additional 2 h. MRE11+/H129N cells were treated with 4-hydroxytamoxifen (4-OHT) for 3 days to inactivate the wild-type MRE11 allele, then treated with etoposide (10μM) for 2 h. Error bars represent standard deviation (SD) of three independent experiments. Asterisk indicates p = 2.8 × 10−2, calculated by Student's t test. Representative images of dot plots are shown in Figure S3E.
(B) Quantification of TOP2ccs in MCF-7 cells with the indicated genotypes relative to the amount of TOP2ccs in wild-type MCF-7 cells. Cells were incubated with serum-free medium for 24 h then treated with etoposide for 2 h. Each experiment was performed independently at least three times. Error bars represent SD. Single, double, triple, and quadruple asterisks indicate p = 4.7 × 10−4, p = 6.4 × 10−3, p = 5.3 × 10−3, and p = 1.9 × 10−2 respectively, calculated by Student's t -test. Representative images of dot plots are shown in Figures S3F and S3G.