Fig. 1 |. Routes for RNA trafficking between plant cells.
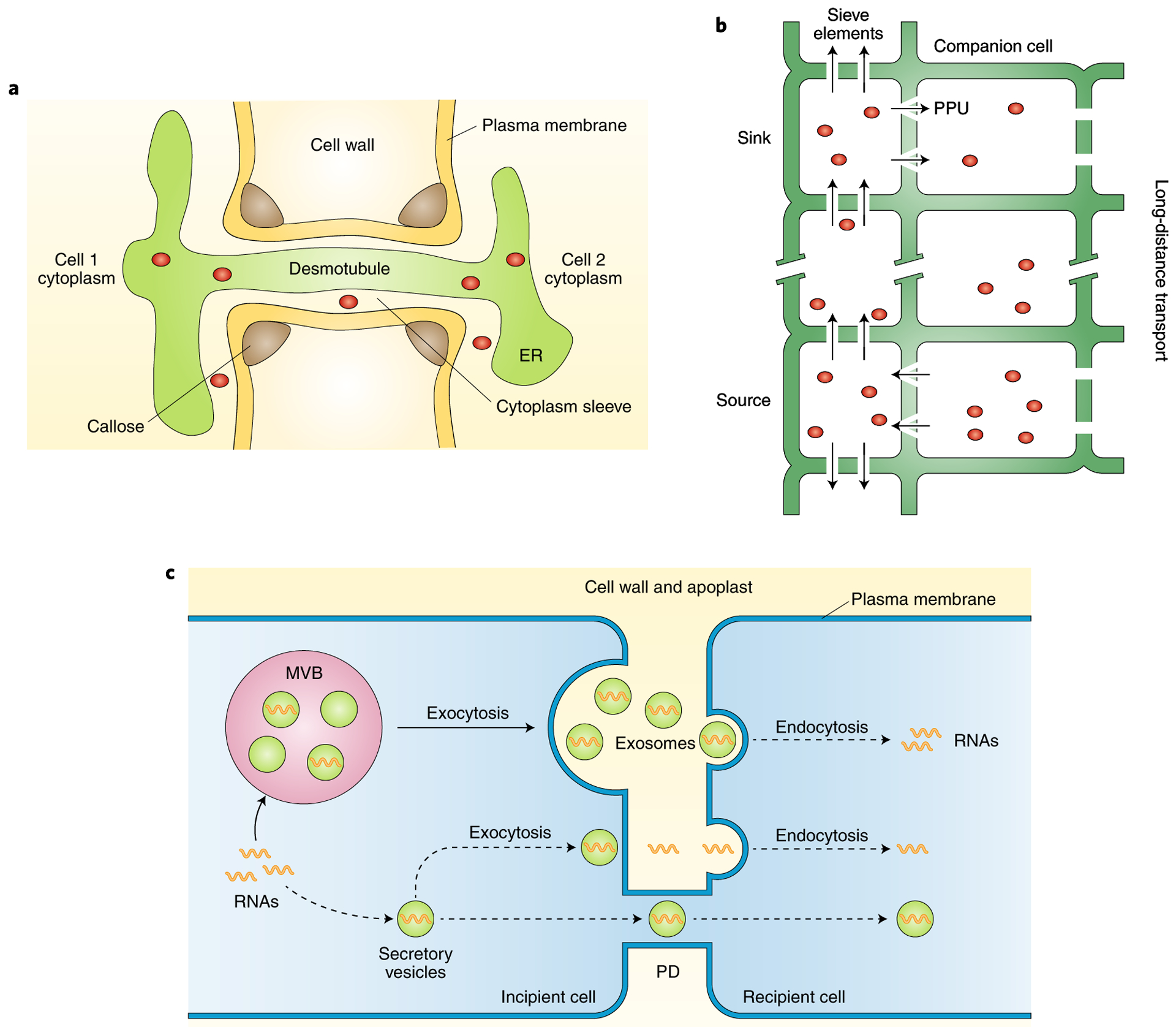
a, PD as micro-channels for cell-to-cell movement of mobile molecules. PD are membrane-lined channels that traverse the cell wall and connect neighbouring cells. Cell wall, plasma membrane, endoplasmic reticulum (ER), desmotubule and callose are indicated. Red circles represent soluble molecules capable of moving through the desmotubule or cytoplasmic sleeve of PD. b, Phloem as a conduit for long-distance movement of molecules. Phloem is composed of stacked enucleated sieve elements assisted by companion cells. Mature sieve elements are connected to adjacent companion cells by highly modified and funnel-like plasmodesmata pore units (PPUs). Mobile molecules (red circles) are primarily transported from source to sink tissues over long distances through phloem. Arrows indicate the direction of movement. Gaps in the line indicate multiple, stacked sieve elements (one cell in the diagram) mediating long-distance transport. c, Putative vesicle-mediated RNA trafficking in plants. Vesicles containing RNAs are taken into MVBs, which subsequently fuse with the plasma membrane and release their intraluminal vesicles into the extracellular space as exosomes. These exosomes fuse with the plasma membrane of the recipient cell through endocytosis and unload the cargo RNAs. Alternatively, vesicles may transport RNAs to adjacent cells, either through exocytosis/endocytosis or through the PD channels between cells. Note that these are purely hypothetical events, as indicated by dashed lines in the diagram.
