Hypercholesterolemia is a strong predictor of cardiovascular diseases. The 3-hydroxy-3-methylglutaryl coenzyme A reductase gene (Hmgcr) coding for the rate-limiting enzyme in the cholesterol biosynthesis pathway is a crucial regulator of plasma cholesterol levels. However, the posttranscriptional regulation of Hmgcr remains poorly understood. The main objective of this study was to explore the role of microRNAs (miRNAs) in the regulation of Hmgcr expression. Systematic in silico predictions and experimental analyses reveal that miRNA 27a (miR-27a) specifically interacts with the Hmgcr 3′ untranslated region in murine and human hepatocytes.
KEYWORDS: hypercholesterolemia, miRNA, Hmgcr, posttranscriptional regulation, cardiovascular disease, cholesterol, gene expression, molecular biology, transcription
ABSTRACT
Hypercholesterolemia is a strong predictor of cardiovascular diseases. The 3-hydroxy-3-methylglutaryl coenzyme A reductase gene (Hmgcr) coding for the rate-limiting enzyme in the cholesterol biosynthesis pathway is a crucial regulator of plasma cholesterol levels. However, the posttranscriptional regulation of Hmgcr remains poorly understood. The main objective of this study was to explore the role of microRNAs (miRNAs) in the regulation of Hmgcr expression. Systematic in silico predictions and experimental analyses reveal that miRNA 27a (miR-27a) specifically interacts with the Hmgcr 3′ untranslated region in murine and human hepatocytes. Moreover, our data show that Hmgcr expression is inversely correlated with miR-27a levels in various cultured cell lines and in human and rodent tissues. Actinomycin D chase assays and relevant experiments demonstrate that miR-27a regulates Hmgcr by translational attenuation followed by mRNA degradation. Early growth response 1 (Egr1) regulates miR-27a expression under basal and cholesterol-modulated conditions. miR-27a augmentation via tail vein injection of miR-27a mimic in high-cholesterol-diet-fed Apoe−/− mice shows downregulation of hepatic Hmgcr and plasma cholesterol levels. Pathway and gene expression analyses show that miR-27a also targets several other genes (apart from Hmgcr) in the cholesterol biosynthesis pathway. Taken together, miR-27a emerges as a key regulator of cholesterol biosynthesis and has therapeutic potential for the clinical management of hypercholesterolemia.
INTRODUCTION
Cardiovascular diseases (CVDs) remain the leading cause of global mortality and morbidity (1). Among various determinants of CVDs, plasma cholesterol is an important factor contributing to multiple disease states, including atherosclerosis, coronary artery disease, obesity, hypertension, and type 2 diabetes (2–4). The 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase gene (human, HMGCR; mouse/rat, Hmgcr), which codes for an ∼97-kDa endoplasmic reticulum (ER) membrane glycoprotein catalyzing the rate-limiting step in the cholesterol biosynthesis pathway (5), is therefore a critical modulator of dyslipidemia and consequent CVDs.
Statins, HMG-CoA reductase inhibitors, are widely used to reduce high cholesterol levels and the risk of CVDs (6). HMGCR expression/enzyme activity is modulated by feedback control mechanisms involving sterols and nonsterols at the transcriptional and posttranslational levels by the family of sterol regulatory element binding proteins (SREBPs), SREBP cleavage-activated protein (SCAP), and insulin-induced genes (Insig1 and Insig2) (7). However, the molecular mechanisms regulating Hmgcr expression at the posttranscriptional level are poorly understood.
MicroRNAs (miRNAs) are small noncoding RNAs that control gene expression by translational repression and/or mRNA degradation. Recent studies have revealed that miRNAs play important roles in cardiovascular physiology and pathophysiology (8, 9). We performed systematic computational and extensive experimental analyses that revealed a crucial role for miRNA 27a (miR-27a) in the posttranscriptional regulation of Hmgcr under basal and elevated-cholesterol conditions. This study also highlights the previously unknown role of early growth response 1 (Egr1) in miR-27a expression. Interestingly, miR-27a targets multiple genes in addition to Hmgcr in the cholesterol biosynthesis pathway. In line with these in vitro findings, miRNA-27a represses Hmgcr expression in liver tissues and modulates plasma cholesterol levels in high-cholesterol diet (HCD)-fed Apoe−/− mice.
RESULTS
Comparative genomic analysis of mouse and rat Hmgcr gene sequences.
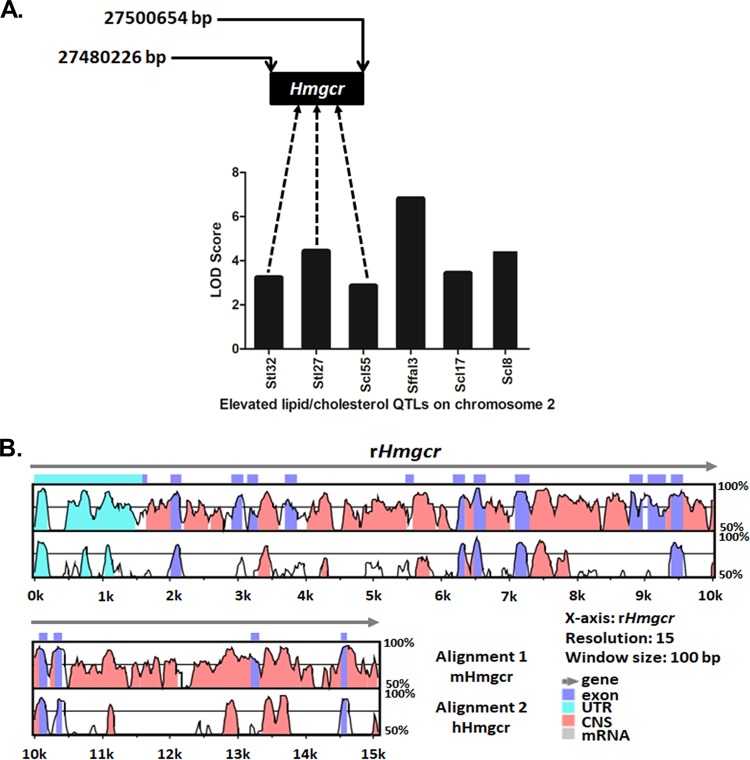
An analysis of rat elevated lipid/cholesterol quantitative trait loci (QTLs) on Hmgcr-harboring chromosome 2 (chr2) within the range of bp 26000000 to 28000000 detected six lipid-related QTLs (Fig. 1A); these QTLs and their respective LOD (logarithm [base 10] of odds) scores were retrieved from the Rat Genome Database (RGD) and are shown in Table 1. Interestingly, among the QTLs harboring the Hmgcr locus (bp 27480226 to 27500654; RGD identifier 2803), Stl27 (bp 23837491 to 149614623) and Stl32 (bp 22612952 to 67612952) displayed significant linkages (LOD scores of 4.4 and 3.2, respectively) with serum triglyceride levels (Fig. 1A), while Scl55 (bp 26186097 to 142053534) showed a significant linkage with serum cholesterol levels (LOD score of 2.83). Moreover, alignment of the mouse and human genomic regions with the orthologous rat sequences at the Hmgcr locus using mVISTA showed >75% homology between the rodent sequences at exons, introns, and untranslated regions (UTRs) (Fig. 1B); the human genomic regions show conservation at the 5′ UTRs and exonic regions with the rat sequences. In general, the extent of homology between each of the 20 Hmgcr exons in mouse and rat was higher (>85%) than that of the noncoding regions (Fig. 1B). Thus, mouse Hmgcr (mHmgcr) appeared to be a logical candidate gene for studying the mechanisms of hypercholesterolemia.
FIG 1.
Graphical representation of rat QTLs contributing to elevated lipid/cholesterol levels and homology between mouse, human, and rat Hmgcr gene sequences. (A) Lipid/cholesterol QTLs and their respective LOD scores (retrieved from the Rat Genome Database). Three of these six QTLs harbor the Hmgcr gene. The genomic position of the rat Hmgcr (rHmgcr) gene is indicated. (B) Conservation analysis of human/mouse and rat Hmgcr sequences using mVISTA. The horizontal axis represents the rHmgcr gene (chr2, bp 27480226 to 27500654) as the reference sequence, whereas the upper vertical axis indicates the percent homology between the rat and mouse Hmgcr genes (chr13, bp 96650579 to 96666685), and the lower vertical axis indicates the percent homology between the rat and human Hmgcr (HMGCR) genes (chr5, bp 75337168 to 75362104). The length of comparison or the window size was set to 100 bp, with a minimum of a 70% match. The annotation of the gene is represented by different colors. The mouse, rat, and human Hmgcr genes are comprised of 20 exons; the 3′ UTRs (untranslated regions) are not visible in the homology plot due to their very small sizes. CNS, conserved noncoding sequences.
TABLE 1.
Genomic positions and LOD scores of various lipid/cholesterol QTLs present on rat chromosome 2 (bp 26000000 to 28000000)a
| Lipid/cholesterol QTL | Start position (bp) | Stop position (bp) | LOD score |
|---|---|---|---|
| Stl32 | 22612952 | 67612952 | 3.2 |
| Stl27 | 23837491 | 149614623 | 4.4 |
| Scl55 | 26186097 | 142053534 | 2.83 |
| Sffal3 | 27760301 | 72760301 | 6.78 |
| Scl17 | 228712271 | 266435125 | 3.4 |
| Scl8 | 231621666 | 266435125 | 4.4 |
These data were retrieved from the Rat Genome Database (http://rgd.mcw.edu/rgdweb/search/qtls.html?100).
Identification of potential miRNAs involved in Hmgcr regulation.
Since miRNA prediction tools employ different algorithms, their outputs may vary, and often, a set of miRNAs predicted by one tool may not overlap those of the others. Hence, in this study, we used multiple tools to predict the miRNA binding sites in the 3′ UTR of mHmgcr to increase the accuracy of target predictions. In silico predictions followed by extensive screening procedures shortlisted 7 miRNAs (miR-27a, miR-27b, miR-28, miR-124, miR-345, miR-351, and miR-708) (Table 2). Since the miR-27a, miR-27b, miR-28, and miR-708 binding sites are highly conserved across mammals, including humans, we examined their expression profiles in different human tissues using the DASHR database. The HMGCR transcript levels in different human tissues were mined from the GTEx portal. Interestingly, HMGCR expression showed a significant inverse correlation with miR-27a expression (Fig. 2A) (Pearson r = −0.9007; P < 0.05). In contrast, miR-27b, miR-28, and miR-708 levels did not exhibit an inverse correlation with HMGCR expression in these tissues (data not shown). Since the expression levels of miR-124, miR-345, and miR-351 across various tissues were near or below the detectable range for the functional repression of target genes (viz., below 100 reads per million [RPM]) (10), these were not considered for correlation analysis. Moreover, only miR-27a and miR-27b have been validated to interact with Hmgcr via high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP) analysis as reported in the TarBase database (Table 3). All these lines of evidence indicate a possible role of miR-27a and miR-27b in the posttranscriptional regulation of Hmgcr. Hence, miR-27a and miR-27b were selected for further investigation of their interactions with Hmgcr.
TABLE 2.
Predicted miRNAs with potential binding sites in the 3′ UTR of mHmgcra
| miRNA | No. of bases in the seed sequence | PITA ΔΔG | RNAhybrid ΔG (kcal/mol) |
|---|---|---|---|
| mmu-miR-124 | 7 | −10.62 | −29.5 |
| mmu-miR-28 | 8 | −10.28 | −22.2 |
| mmu-miR-345-5p | 8 | −11.08 | −32 |
| mmu-miR-351 | 8 | −16.89 | −33.6 |
| mmu-miR-708 | 7 | −14.34 | −27.4 |
| mmu-miR-27a | 6 | −10.13 | −20.7 |
| mmu-miR-27b | 6 | −10.74 | −22.6 |
miRNAs predicted by at least 5 programs that displayed a PITA ΔΔG score of less than −10 and an RNAhybrid ΔG value of −20 kcal/mol were selected.
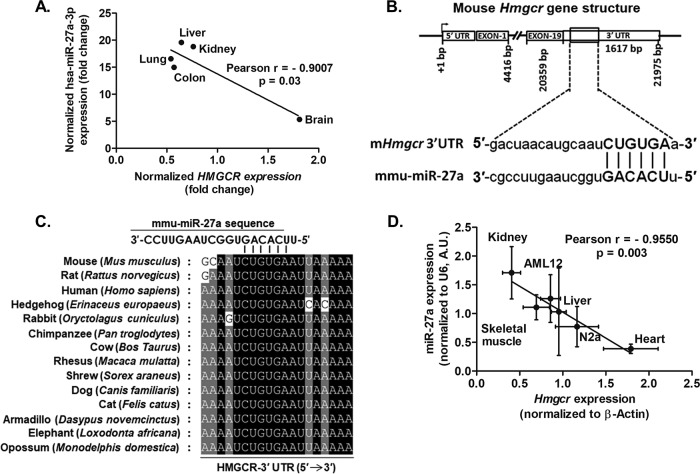
FIG 2.
miR-27a binding sites in the 3′ UTR of Hmgcr and inverse correlation between mmu-miR-27a-3p/hsa-miR-27a-3p and Hmgcr/HMGCR expression. (A) Inverse correlation between HMGCR and hsa-miR-27a-3p expression in different human tissues. (B) Schematic representation of Hmgcr showing the miR-27a binding site in the 3′ UTR. The complementarity between the seed region of miR-27a and the mHmgcr 3′ UTR is depicted in capital letters. (C) Conservation of the miR-27a binding site in the 3′ UTR of Hmgcr across different mammals. (D) Negative correlation between Hmgcr mRNA and miR-27a expression in cultured AML12 cells, N2a cells, and rat tissues (the number of biological/technical replicates was at least 3 in each case). A.U., arbitrary units.
TABLE 3.
In silico tools and databases employed in the study
Direct interaction of miR-27a with the Hmgcr 3′ UTR downregulates Hmgcr protein levels in hepatocytes.
Watson-Crick base pairing between miR-27a and the Hmgcr 3′ UTR revealed that the 2- to 7-base seed sequence of miR-27a has perfect complementarity to the mHmgcr 3′ UTR (Fig. 2B). Since the miR-27a binding site is highly conserved across mammals (Fig. 2C), a significant negative correlation between miR-27a and Hmgcr expression was observed in mouse hepatocyte and neuroblast cells and in rat liver, kidney, and skeletal muscle tissues (Pearson r = −0.9550; P < 0.05) (Fig. 2D).
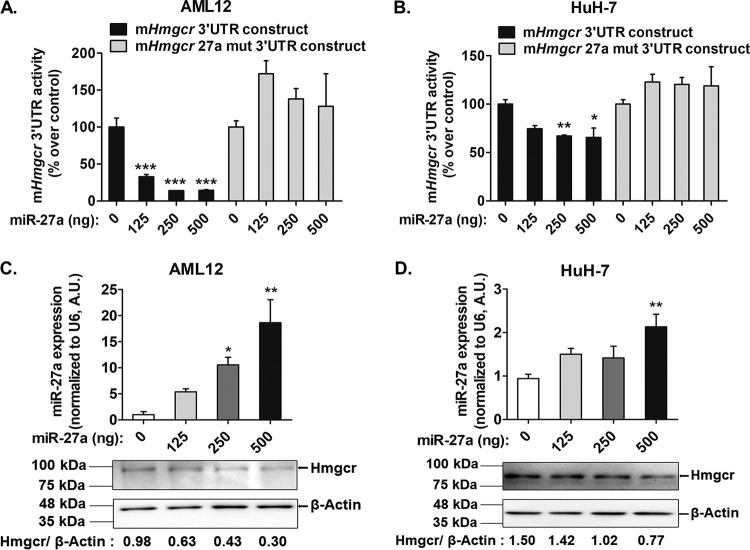
To examine whether miR-27a directly interacts with Hmgcr, an miR-27a expression plasmid was cotransfected with mHmgcr 3′-UTR–luciferase construct into AML12 (alpha mouse liver 12) and HuH-7 cells (Fig. 3A and B). Indeed, miR-27a overexpression caused a significant dose-dependent reduction in the mHmgcr 3′-UTR reporter activity in both AML12 (up to ∼85%; P < 0.001) and HuH-7 (up to ∼34%; P < 0.01) cells. In contrast, cotransfection of the mHmgcr 27a mut 3′-UTR construct (devoid of the miR-27a binding site) with the miR-27a expression plasmid showed no significant change in 3′-UTR reporter activity (Fig. 3A and B). Furthermore, quantitative real-time PCR (qPCR) analysis showed that the overexpression of pre-miR-27a led to a dose-dependent increase in miR-27a expression in AML12 (up to ∼1,770%; P < 0.01) and HuH-7 (up to ∼125%; P < 0.01) cells. Consistent with the mHmgcr 3′-UTR reporter activity, the overexpression of miR-27a caused a decrease in the endogenous Hmgcr/HMGCR protein level (Fig. 3C and D).
FIG 3.
miR-27a negatively regulates Hmgcr expression in cultured hepatocytes. (A and B) The mHmgcr 3′-UTR reporter construct or the mHmgcr 27a mut 3′-UTR construct (500 ng) was cotransfected with the miR-27a expression plasmid in AML12 (A) and HuH-7 (B) cells, followed by luciferase assays (n = 5). The results are means ± SEM of triplicate values. (C and D) Transfection of the miR-27a expression plasmid results in overexpression of miR-27a in AML12 (C) and HuH-7 (D) cells (n = 5). Statistical significance was determined by one-way ANOVA with a Newman-Keuls multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared to the control). Hmgcr downregulation was confirmed by Western blotting (C and D) (n = 3).
Despite the predicted binding site of miR-27b in the Hmgcr 3′-UTR, cotransfection of the miR-27b expression plasmid did not cause a significant decrease in mHmgcr 3′-UTR luciferase reporter activity (data not shown). Moreover, to test the specificity of interactions of miR-27a with the Hmgcr 3′-UTR, we carried out cotransfection experiments with miR-764, which does not have binding sites in the Hmgcr 3′-UTR; no change in Hmgcr 3′-UTR luciferase reporter activity was observed (data not shown).
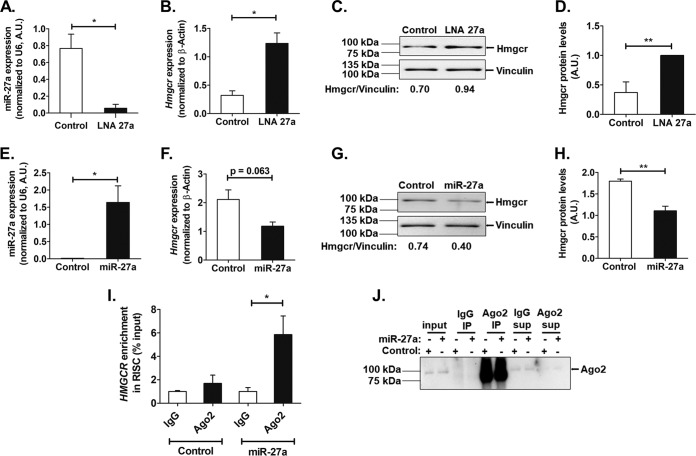
Furthermore, transfection of a locked nucleic acid inhibitor of miR-27a (LNA 27a) in AML12 cells diminished endogenous miR-27a levels (by ∼15-fold; P < 0.05) (Fig. 4A) and enhanced Hmgcr mRNA (by ∼3.8-fold; P < 0.05) (Fig. 4B) and protein (Fig. 4C and D) levels. On the other hand, transfection of an miR-27a mimic showed enhanced miR-27a levels (by ∼164-fold; P < 0.05) (Fig. 4E) and diminished Hmgcr mRNA (Fig. 4F) and protein (Fig. 4G and H) levels. In corroboration, RIP (ribonucleoprotein immunoprecipitation) assays with an antibody specific for Ago2 (an integral component of the RNA-induced silencing complex [RISC]) showed an enrichment of the HMGCR transcript level (by ∼3.5-fold; P < 0.05) in the Ago2-immunoprecipitated RNA fraction of HuH-7 cells overexpressing miR-27a compared to the control conditions, thereby confirming the interaction of miR-27a with the Hmgcr 3′-UTR in the context of the RISC (Fig. 4I). The RNA fraction immunoprecipitated using preimmune anti-mouse IgG antibody (control) showed no significant difference in HMGCR levels between miR-27a overexpression and basal conditions (Fig. 4I). Pulldown of Ago2 was confirmed by Western blotting (Fig. 4J).
FIG 4.
Specific interactions of miR-27a with the Hmgcr 3′-UTR. (A and B) Relative expression levels of miR-27a (A) and Hmgcr (B) transcripts upon transfection of 60 nM the control oligonucleotide or locked nucleic acid inhibitor of miR-27a (LNA 27a) in AML12 cells were determined by qPCR (n = 3). (C and D) Representative Western blot analysis of Hmgcr protein levels in AML12 cells upon transfection of 60 nM the control oligonucleotide or LNA27a (n = 3) (C) along with densitometric analysis of Hmgcr protein levels normalized to vinculin levels (D). The bar plot shown is representative of data from three experiments. (E and F) miR-27a (E) and Hmgcr (F) transcript levels upon transfection of 1 μg of the control oligonucleotide or the miR-27a mimic in AML12 cells were determined by qPCR (n = 3). (G and H) Representative Western blot analysis of Hmgcr protein levels in AML12 cells upon transfection of 1 μg of the control oligonucleotide or the miR-27a mimic (n = 3) (G) along with densitometric analysis of Hmgcr protein levels normalized to vinculin levels (H). The bar plot shown is representative of data from three experiments. Statistical significance was determined by Student’s t test (unpaired, 2 tailed). *, P < 0.05; **, P < 0.01 (compared to the control). (I) Ago2 ribonucleoprotein precipitation analysis in HuH-7 cells overexpressing miR-27a. HMGCR enrichment was normalized to the corresponding input under each condition. Data are expressed as a percentage of the input and are the means ± SEM for triplicates (n = 3). (J) Ago2 immunoprecipitation (IP) was confirmed by Western blotting (n = 3). Statistical significance was determined by one-way ANOVA with a Newman-Keuls multiple-comparison test. *, P < 0.05 (with respect to control Ago2 conditions). sup, supernatant.
miR-27a represses Hmgcr by posttranslational inhibition followed by mRNA degradation.
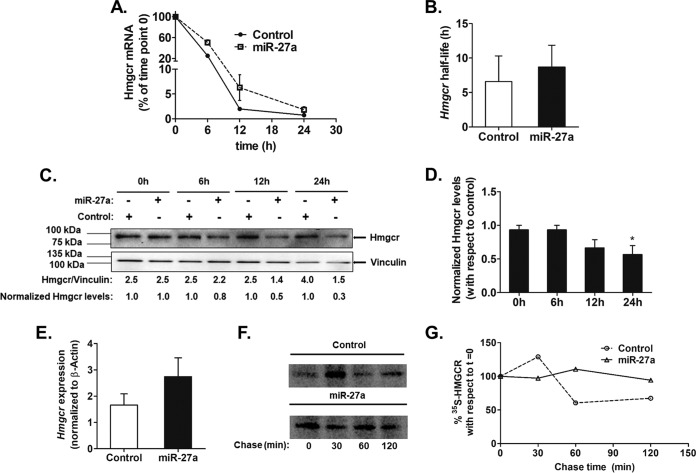
In order to unfold the mechanism of action of miR-27a on Hmgcr, mRNA stability assays using actinomycin D were performed in AML12 cells overexpressing miR-27a (Fig. 5). Interestingly, no significant change in the Hmgcr mRNA half-life (t1/2) was observed upon miR-27a overexpression until 24 h (Fig. 5A and B). In contrast, Hmgcr protein levels showed a time-dependent reduction following actinomycin D treatment in AML12 cells transfected with miR-27a (Fig. 5C and D). In corroboration, the overexpression of miR-27a did not alter the steady-state Hmgcr mRNA level (Fig. 5E). However, 36 h after the transfection of LNA 27a in AML12 cells, endogenous Hmgcr mRNA levels were significantly enhanced (Fig. 4B). Furthermore, HuH-7 cells transfected with the miR-27a plasmid exhibited no change in 35S-radiolabeled HMGCR level over a chase time of 120 min compared to the HuH-7 cells transfected with pcDNA 3. These observations suggest that Hmgcr repression by miR-27a is mediated predominantly by translational attenuation followed by mRNA degradation (Fig. 5F and G).
FIG 5.
miR-27a regulates Hmgcr expression by translational repression in cultured hepatocytes. AML12 cells were transfected with 500 ng of the miR-27a expression plasmid or pcDNA3.1 (as a control). After 12 h of transfection, they were incubated with actinomycin D (5 μg/ml) for different times. (A) Hmgcr mRNA levels were plotted relative to the 0-h time point as described in Materials and Methods (n = 3). (B) Endogenous Hmgcr mRNA half-life estimation in AML12 cells upon the ectopic overexpression of miR-27a. The mRNA half-life of Hmgcr was measured over 24 h in the presence of 5 μg/ml of actinomycin D in control cells (transfected with pcDNA3.1) and miR-27a-transfected AML12 cells. The Hmgcr mRNA half-lives are presented as means ± SEM of data from three independent experiments. (C and D) Effect of transcriptional attenuation on endogenous Hmgcr protein levels in miR-27a-overexpressing hepatocytes. AML12 cells were transfected with either pcDNA 3.1 or the miR-27a expression plasmid and incubated with actinomycin D for different time points. (C) Western blot analysis of total proteins was carried out to probe for Hmgcr and vinculin. The relative Hmgcr protein levels normalized to vinculin levels at different time points are also shown. The normalized Hmgcr levels as fold changes over the control for every time point of actinomycin D treatment are also indicated (n = 3). (D) Bar plot showing the normalized Hmgcr protein levels expressed as fold changes over the corresponding controls for different time points of actinomycin D treatment from three experiments. (E) Relative expression of Hmgcr 24 h after transfection of the miR-27a expression plasmid in AML12 cells was determined by qPCR using gene-specific primers (n = 3). Hmgcr expression was normalized to β-actin mRNA expression in the same sample. Statistical significance was determined by Student’s t test (unpaired, 2 tailed). *, P < 0.05 (compared to the control). (F) Pulse-chase analysis of HMGCR in HuH-7 cells transfected with the miR-27a expression plasmid. After 24 h of transfection, the cells were labeled with [35S]Met for an hour, followed by a chase up to 120 min. The cells were then lysed, immunoprecipitated with the anti-HMGCR antibody, and analyzed by autoradiography. (G) Densitometric analysis of 35S-labeled HMGCR levels in mock- and miR-27a-transfected lysates.
Role of intracellular cholesterol levels in miR-27a-mediated regulation of Hmgcr.
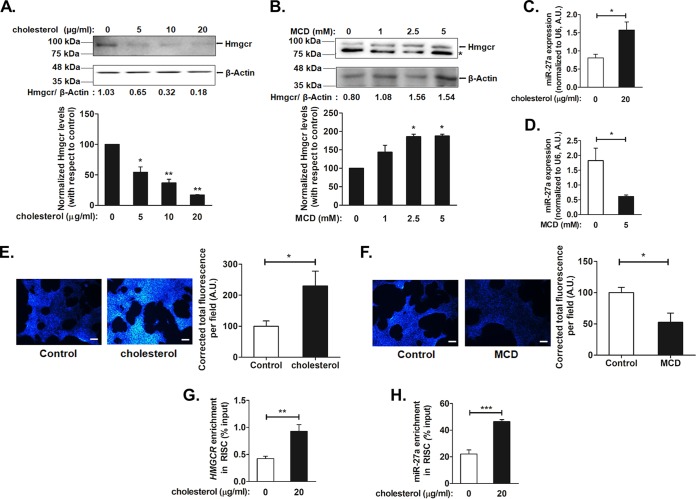
Because the Hmgcr protein level is dependent on the intracellular cholesterol level and regulated by a negative feedback mechanism, we sought to determine whether modulating the endogenous cholesterol level affects the posttranscriptional regulation of Hmgcr. Accordingly, AML12 cells were treated with different concentrations of either cholesterol (5, 10, and 20 μg/ml) or methyl-β-cyclodextrin (MCD) (an oligosaccharide that reduces intracellular cholesterol levels) (1, 2.5, and 5 mM), followed by Western blotting for Hmgcr (Fig. 6A and B). Since 20 μg/ml of cholesterol and 5 mM MCD showed effective reduction and augmentation of the Hmgcr protein level, respectively, these doses were used for further experiments. Interestingly, cholesterol treatment showed ∼1.9-fold (P < 0.05) enhancement of the miR-27a level (Fig. 6C), while cholesterol depletion caused ∼3-fold (P < 0.05) reduction in the endogenous miR-27a level (Fig. 6D). The increase or decrease in the intracellular cholesterol level was confirmed by filipin staining (Fig. 6E and F). Furthermore, Ago2-RIP assays of cholesterol-treated HuH-7 cells revealed significant enrichments of HMGCR (∼2.2-fold; P < 0.01) (Fig. 6G) and miR-27a (∼1.7-fold; P < 0.001) (Fig. 6H) levels, suggesting an interaction of HMGCR with miR-27a under elevated-cholesterol conditions.
FIG 6.
Intracellular cholesterol modulates the expression of miR-27a and augments the interactions of miR-27a with the Hmgcr 3′ UTR. AML12 cells were treated with increasing doses of either cholesterol or the cholesterol-depleting reagent methyl-β-cyclodextrin (MCD) for 6 h or 15 min, respectively. After incubation for 6 to 9 h in serum-free medium, the cells were processed for RNA and protein isolation and fluorescence microscopy. (A and B) Western blot analysis of total protein isolated from AML12 cells treated with cholesterol (A) or MCD (B) was carried out for Hmgcr and β-actin levels (n = 3). * indicates a nonspecific band. Hmgcr protein levels from independent experiments were normalized to β-actin levels and are shown in the bar plot below the representative Western blots. Statistical significance was determined by one-way ANOVA with Bonferroni’s multiple-comparison test. *, P < 0.05; **, P < 0.01 (compared to basal conditions). (C and D) miR-27a levels were also determined by qPCR upon exogenous cholesterol (20 μg/ml) (C) or MCD (5 mM) (D) treatment in AML12 cells (n = 3). The miR-27a expression levels were normalized to U6 RNA levels and are indicated as means ± SEM for triplicate values. (E and F) Cholesterol (E) or MCD (F) treatments in AML12 cells were confirmed by staining intracellular cholesterol using filipin stain followed by fluorescence microscopy. Bars, 170 μm. Quantification of the total corrected fluorescence per field was performed after cholesterol or MCD treatment for four experiments using ImageJ and is represented as a bar plot beside the representative image. Statistical significance was determined by Student’s t test (unpaired, 2 tailed). *, P < 0.05 (compared to control conditions). (G and H) Enrichment of HMGCR and miR-27a upon exogenous cholesterol treatment in Ago2-immunoprecipitated RNA from HuH-7 cells. HuH-7 cells were treated with 20 μg/ml of cholesterol for 6 h, and the total RNA fraction from the Ago2/IgG-immunoprecipitated samples (in basal and cholesterol-treated cells) was subjected to qPCR using HMGCR (G)- and miR-27a (H)-specific primers (Table 4) (n = 3). The HMGCR or miR-27a enrichment was normalized to the corresponding input under each condition and is represented as a percentage of the input. Statistical significance was determined by Student’s t test (unpaired, 2 tailed). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared to basal conditions).
Role of Egr1 in miR-27a expression under basal and modulated-intracellular-cholesterol conditions.
To understand the possible mechanism of miR-27a regulation, we predicted transcription factor binding sites in the miR-27a promoter domain (∼500 bp) using two programs: LASAGNA and JASPAR (Table 3). Egr1, a transcription factor (predominantly expressed in the liver) that plays a crucial role in the transcriptional regulation of most cholesterol biosynthesis genes, including Hmgcr (11), had six putative binding sites as two separate clusters in the miR-27a promoter domain (Fig. 7A).
FIG 7.
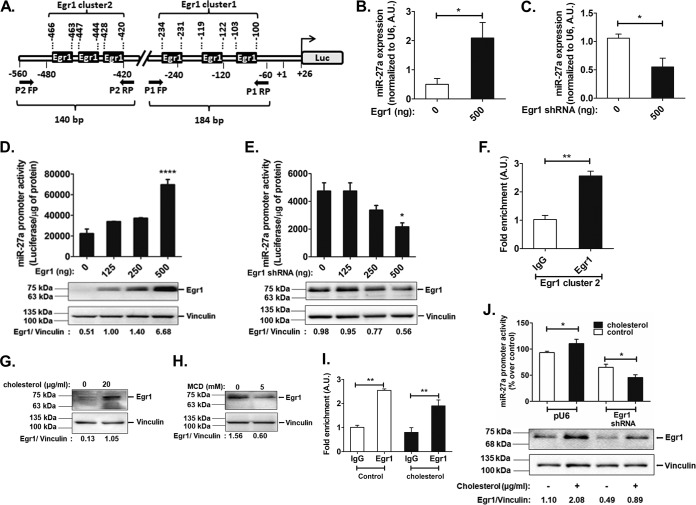
Egr1 modulates miR-27a expression under basal and altered-cholesterol conditions. (A) Schematic representation of the proximal mmu-miR-27a promoter domain harboring multiple Egr1 binding sites. (B and C) AML12 cells were cotransfected with the mmu-miR-27a promoter (500 ng) and 500 ng of either the Egr1 expression plasmid (B) or the Egr1 shRNA expression plasmid (C), followed by qPCR to probe for the endogenous levels of miR-27a (normalized to U6 RNA levels) (n = 3). Statistical significance was determined by Student’s t test (unpaired, 2 tailed). *, P < 0.05 (compared to control conditions). (D and E) AML12 cells were cotransfected with increasing doses of either the Egr1 expression plasmid (D) or the Egr1 shRNA expression plasmid (E) and the miR-27a promoter construct (500 ng) (n = 5), followed by luciferase assays. Results are expressed as means ± SEM for triplicate values. Statistical significance was determined by one-way ANOVA with a Newman-Keuls multiple-comparison test. *, P < 0.05; ****, P < 0.0001 (compared to basal conditions). The overexpression or downregulation of Egr1 was confirmed by Western blotting (n = 3). (F) ChIP assays using Egr1/IgG-immunoprecipitated chromatin isolated from AML12 cells. qPCR was performed with immunoprecipitated DNA using the P2 primer pair (amplifying Egr1 cluster 2) in the miR-27a promoter (n = 3). Statistical significance was determined by Student’s t test (unpaired, 2 tailed). **, P < 0.01 (compared to the control). (G and H) AML12 cells were treated with either 20 μg/ml of cholesterol [(3β)-cholest-5-en-3-ol] (G) or 5 mM the cholesterol-depleting reagent methyl-β-cyclodextrin (MCD) (H) for 6 h or 15 min, respectively. Western blots quantifying Egr1 levels are shown (n = 3). (I) ChIP assays using Egr1/IgG-immunoprecipitated chromatin isolated from AML12 cells treated with cholesterol (20 μg/ml) for 6 h. Statistical significance was determined by one-way ANOVA with Bonferroni’s multiple-comparison posttest. **, P < 0.01 (compared to the corresponding IgG). (J) AML12 cells were cotransfected with either pU6 (empty vector as a control) or the Egr1 shRNA expression plasmid and the miR-27a promoter construct (500 ng) (n = 6). After 24 h of transfection, the cells were treated with cholesterol (20 μg/ml) for 6 h and lysed for luciferase and Bradford assays. The luciferase activity was normalized to the total protein and expressed as a percentage over the pU6 control. The results are expressed as means ± SEM. Statistical significance was determined by one-way ANOVA with a Newman-Keuls multiple-comparison test. *, P < 0.05 (compared to the respective control conditions). Western blot analysis was performed to probe for Egr1 levels upon the transfection of AML12 cells with pU6 or the Egr1 shRNA plasmid, followed by cholesterol treatment. The Egr1 protein levels were normalized to vinculin levels and are indicated below the representative Western blot image. The blot shown is representative of results from three experiments.
Next, we validated the role of Egr1 in miR-27a expression by cotransfection experiments. A concomitant increase (∼4-fold; P < 0.05) and decrease (∼2-fold; P < 0.05) in endogenous miR-27a levels upon Egr1 overexpression and downregulation, respectively, were observed (Fig. 7B and C). Consistently, Egr1 overexpression caused ∼3-fold enhancement of miR-27a promoter activity (P < 0.0001) (Fig. 7D), while Egr1 downregulation resulted in ∼2.2-fold reduction in miR-27a promoter activity (P < 0.05) (Fig. 7E). Thus, Egr1 may play a crucial role in the transcriptional activation of miR-27a. Indeed, ChIP (chromatin immunoprecipitation) assays confirmed the in vivo interaction of Egr1 with the miR-27a-proximal (∼500-bp) promoter domain. qPCR analysis using Egr1 antibody-immunoprecipitated chromatin revealed ∼2.5-fold enrichment of the miR-27a promoter domain with primer pairs for cluster 2 (P < 0.001) (Fig. 7F), whereas no significant fold enrichment of the miR-27a promoter domain was observed using the primer pair for cluster 1 (data not shown), suggesting that the Egr1 sites predicted in this domain (Egr1 cluster 1) may not be functional under basal conditions. Interestingly, Western blot analysis revealed that exogenous cholesterol treatment augmented the Egr1 level (Fig. 7G), while its level diminished upon cholesterol depletion (Fig. 7H), suggesting that the intracellular cholesterol level may regulate miR-27a expression via Egr1.
In order to test if exogenous cholesterol modulates the binding of Egr1 to the miR-27a promoter domain, ChIP assays were performed in AML12 cells treated with cholesterol. However, there was no change in miR-27a promoter occupancy at Egr1 cluster 2 under excess-cholesterol conditions (Fig. 7I). Furthermore, the downregulation of Egr1 abrogated the cholesterol-mediated activation of miR-27a promoter activity (Fig. 7J), suggesting that Egr1 plays a key role in regulating miR-27a expression under conditions of modulated cholesterol levels.
miR-27a mimic reduces hepatic Hmgcr expression and plasma cholesterol levels in high-cholesterol-diet-fed Apoe−/− mice.
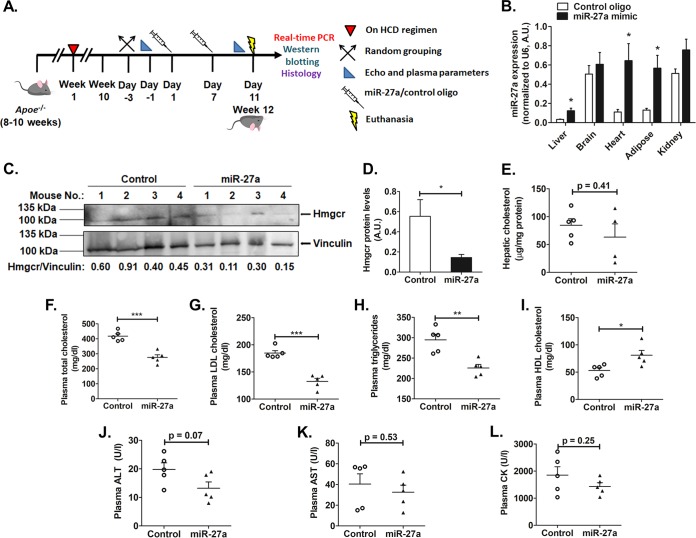
To test if miR-27a mimic can modulate plasma lipid levels in a model of hyperlipidemia, Apoe−/− mice on a high-cholesterol-diet regimen for 10 weeks were injected with either saline or 5 mg/kg of body weight of the miR-27a mimic or a control oligonucleotide as lipid emulsion formulations (Fig. 8A). Tissue analysis of miR-27a levels revealed ∼4-fold (P < 0.05), ∼5.8-fold (P < 0.05), and ∼4.3-fold (P < 0.05) upregulations of miR-27a expression in the liver, heart, and adipose tissues, respectively, in miR-27a mimic-injected animals compared to the control oligonucleotide group. However, miR-27a levels did not differ in the kidney and brain tissues of the miR-27a mimic group compared to the control group (Fig. 8B), suggesting the efficient delivery of the miR-27a mimic to the liver, heart, and adipose tissues.
FIG 8.
miR-27a diminishes Hmgcr and plasma lipid levels in high-cholesterol-diet-fed Apoe−/− mice. (A) Male Apoe−/− mice (8 to 10 weeks old) were fed a high-cholesterol diet (HCD) regimen for 10 weeks, injected with either the miR-27a mimic or the control oligonucleotide twice over a period of 11 days, and then euthanized. (B) qPCR for miR-27a levels in various tissues. (C) Western blot analysis of Hmgcr protein in Apoe−/− mouse liver tissues (n = 4 in each group). Data are representative of results from three experiments. (D) Hmgcr protein levels were normalized to vinculin levels (shown in the bar plot). The blot shown is representative of results from three experiments. (E) Lipids were extracted from liver tissue, and hepatic cholesterol levels were measured in control oligonucleotide- and miR-27a-injected Apoe−/− mice. (F to I) Plasma levels of total cholesterol (F), LDL cholesterol (G), triglycerides (H), and HDL cholesterol (I) from animals fasted overnight. (J to L) Plasma ALT (J), AST (K), and CK (L) levels were also measured. Data are means ± SEM (n = 5 to 6 animals per group). U/l, units per liter. Statistical significance was determined by Student’s t test (unpaired, 2 tailed). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared to the control group).
Western blot analysis of liver tissues of these mice revealed diminished Hmgcr protein levels in miR-27a mimic-injected mice compared to controls (Fig. 8C and D). The anti-Hmgcr antibody detected an ∼120-kDa band, suggesting a predominantly glycosylated form of the protein in these tissues. The hepatic cholesterol level in miR-27a-injected animals showed a modest decrease, which did not differ significantly in comparison to the control group (Fig. 8E).
Next, we assessed the effect of miR-27a mimics on the circulating lipid levels. These parameters were measured 4 days after the second injection of the miR-27a mimic or control oligonucleotide. Interestingly, the miR-27a mimic group showed reduced plasma total cholesterol (∼1.5-fold; P < 0.001), low-density lipoprotein (LDL) cholesterol (∼1.4-fold; P < 0.001), and triglyceride (∼1.3-fold; P < 0.01) (Fig. 8F to H) levels compared to the control group. In contrast, the miR-27a mimic enhanced plasma high-density lipoprotein (HDL) cholesterol levels (∼1.5-fold; P < 0.05) (Fig. 8I) in comparison to the control group.
It is important to note that plasma alanine aminotransferase (ALT), aspartate aminotransferase (AST), and creatine kinase (CK) levels were not elevated in this model, indicating no obvious liver or muscle injury upon miR-27a mimic injections (Fig. 8J to L). These findings suggest the therapeutic potential of an miR-27a mimic as a safe and efficient cholesterol-lowering agent.
miR-27a targets multiple genes in the cholesterol regulatory pathways.
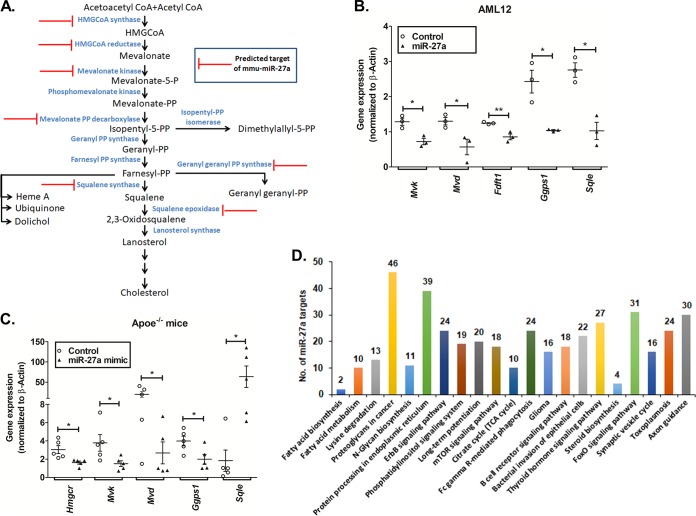
In line with our observations and the reported role of miR-27a in lipid metabolism, we tested if miR-27a targets other genes crucial for cholesterol homeostasis. Interestingly, in silico predictions and PANTHER pathway analysis revealed six additional cholesterol biosynthesis-related gene targets: 3-hydroxy-3-methylglutaryl CoA synthase 1 (Hmgcs1), mevalonate kinase (Mvk), diphosphomevalonate decarboxylase (Mvd), geranylgeranyl pyrophosphate synthase (Ggps1), squalene synthase (Fdft1), and squalene epoxidase (Sqle) (Fig. 9A). Indeed, miR-27a mimic treatment of AML12 cells or high-cholesterol-diet-fed Apoe−/− mice diminished the expression of Mvk, Fdft1, Sqle, Ggps1, and Mvd (Fig. 9B and C). In addition, miR-27a augmentation also resulted in the diminished expression of lipoprotein uptake-related genes (viz., low-density lipoprotein receptor [Ldlr] and scavenger receptor class B member 1 [Scarb1]) (data not shown). Thus, miR-27a seems to play a key role in the global regulation of genes involved in cholesterol homeostasis.
FIG 9.
Augmentation of miR-27a represses multiple genes in the cholesterol biosynthesis pathway. (A) Potential targets of miR-27a in the cholesterol biosynthesis pathway predicted by miRWalk, RNAhybrid, and TargetScan were categorized based on their molecular functions using the PANTHER database. (B and C) qPCR analysis of predicted genes in AML12 cells (n = 3) (B) and liver tissues of high-cholesterol-diet-fed Apoe−/− mice (C) transfected or injected with the miR-27a mimic or the control oligonucleotide (n = 5 to 6 animals per group). Statistical significance was determined by Student’s t test (unpaired, 2 tailed). *, P < 0.05; **, P < 0.01 (compared to the control group). (D) Pathway analysis for miR-27a targets was performed using TargetScan (in silico predictions) and mirPath v3 (validated interactions). Twenty-one pathways were commonly enriched by both these tools, including steroid biosynthesis, fatty acid metabolism, and biosynthesis. This analysis further demonstrates that miR-27a regulates lipid metabolism. The number of genes targeted by miR-27a in each of these pathways is indicated above each bar. TCA, tricarboxylic acid.
DISCUSSION
Overview.
In view of the important role of Hmgcr in cholesterol biosynthesis, its potential regulation by microRNAs has been of major interest in recent years. Notably, miR-29a, miR-185, miR-150, miR-548p, miR-21, miR-195, and miR-342 have so far been reported to interact with the Hmgcr transcript (12–18). We undertook extensive computational and experimental analyses to identify the key miRNAs that may regulate Hmgcr gene expression under basal as well as pathophysiological conditions. Our in vitro data provide several lines of evidence for the regulation of Hmgcr expression by miR-27a (Fig. 2 to 4). This is consistent with a previous study demonstrating the repression of exogenous Hmgcr 3′-UTR activity by miR-27a mimics in hepatocytes (19). However, the regulation of endogenous Hmgcr protein levels in liver cells and the mechanisms of miR-27a-mediated repression of Hmgcr expression were unknown. We also observed that the miR-27a mimic downregulated the hepatic expression of Hmgcr and plasma lipid levels in a high-cholesterol-diet-fed atherosclerotic mouse model (Fig. 8).
Pathophysiological implications of Hmgcr regulation by miR-27a.
HMGCR is tightly regulated by sterols via transcriptional, posttranscriptional, and posttranslational mechanisms (20). In brief, elevated levels of sterols diminish HMGCR expression by inhibiting the sterol regulatory element binding protein 2 (SREBP-2) transcription factor (21). The posttranscriptional and posttranslational regulatory systems operate independently of the SREBP pathway and form an important aspect of sterol-mediated HMGCR regulation. Posttranslational regulation is executed by sterol or nonsterol intermediates via an INSIG-dependent ER-associated protein degradation (ERAD) mechanism involving the ubiquitin-proteasomal degradation of HMGCR (7). However, the effect of elevated sterols on miRNA-mediated HMGCR regulation is partially understood. In view of the crucial role of sterols in HMGCR regulation, do enhanced cholesterol levels modulate endogenous miR-27a levels? Indeed, cholesterol treatment of AML12 cells enhanced miR-27a levels and diminished Hmgcr protein levels (Fig. 6A and C). In order to rule out the possibility that this repression of the Hmgcr protein level is solely because of the INSIG-mediated ERAD mechanism, we performed RIP assays in HuH-7 cells treated with cholesterol. Our Ago2-RIP assays further confirmed the enhanced interaction of miR-27a with HMGCR under conditions of elevated cholesterol levels, suggesting that the posttranscriptional regulation of Hmgcr by miR-27a is an additional mechanism for Hmgcr repression under high-cholesterol conditions (Fig. 6G and H). Thus, this study revealed the crucial role of intracellular cholesterol in Hmgcr expression via miR-27a. This is in corroboration with the enhanced miR-27a levels in liver tissues of Apoe−/− mice fed a high-cholesterol diet compared to Apoe−/− mice fed with a normal chow diet (data not shown).
Are there other microRNAs that modulate lipid or lipoprotein synthesis/secretion in the circulation? There are limited reports on the regulatory roles of miRNAs in cholesterol homeostasis. miR-122 inhibition in mice results in diminished plasma cholesterol and triglyceride levels, with reduced fatty acid and cholesterol synthesis (22). Interestingly, the deletion of miR-122 in mice causes enhanced hepatic lipid, plasma alanine aminotransferase, and alkaline phosphatase levels, with the development of steatohepatitis, fibrosis, and hepatocellular carcinoma (HCC) with age (23). Likewise, miR-34 overexpression in mice reduced plasma triglyceride and cholesterol levels but augmented hepatic triglyceride levels, causing hepatosteatosis (24). Recent studies indicate that miR-30c lowers plasma cholesterol levels in different mouse models of atherosclerosis, hypercholesterolemia, and metabolic disorders without increasing plasma transaminase levels or causing hepatic steatosis (25, 26). These studies highlight the potential of miRNAs as therapeutic agents in the treatment of hypercholesterolemia.
For validation of our in vitro findings, we tested the effect of the miR-27a mimic using high-cholesterol-diet-fed Apoe−/− mice. An improved lipid-based formulation was employed for the efficient delivery of the miR-27a mimic via tail vein injection to major organs (viz., the liver, heart, and adipose tissues, as confirmed by tissue analysis of miR-27a levels) (Fig. 8B). The administration of miR-27a over a period of 11 days induced a significant reduction in the plasma total cholesterol, LDL cholesterol, and triglyceride levels, with a concomitant increase in HDL cholesterol levels (Fig. 8F to I).
miR-27a appears to diminish plasma cholesterol levels by reducing the Hmgcr protein level in the liver (Fig. 8C and D). The reductions in plasma triglyceride levels are consistent with a previous report wherein miR-27a expression diminished triglyceride levels in mouse hepatocytes mainly by targeting fatty acid synthase (Fas) and stearyl CoA desaturase 1 (Scd1) (27). We speculate that the increase in the plasma HDL cholesterol level might result from the miR-27a-mediated regulation of Scarb1, an HDL cholesterol receptor (data not shown). It is interesting to note that food-derived remnant cholesterol has a modest contribution to the total cholesterol level. Moreover, it was previously reported that remnant lipoproteinemia in APOE−/− minipigs is not efficient in initiating atherosclerosis but contributes to the atherosclerotic progression of preexisting lesions (28). Therefore, the miR-27a mimic may diminish overall plasma total cholesterol levels in high-cholesterol-diet-fed Apoe−/− mice by modulating LDL, HDL, and other cholesterol fractions.
Does miR-27a mimic treatment cause any adverse effects in the diet-induced atherosclerosis mouse model? We did not detect any significant changes in plasma ALT, AST, and CK levels, suggesting that the animals did not suffer any liver or muscle injury (Fig. 8J to L). However, additional studies are required to test the long-term effects of the miR-27a mimic to further evaluate its therapeutic potential.
Molecular mechanisms of Hmgcr regulation by miR-27a.
Generally, miRNAs exert their action by translational inhibition followed by mRNA deadenylation, decapping, and decay (29). Our actinomycin D chase experiments showed that the Hmgcr mRNA half-life did not change, whereas the Hmgcr protein level diminished in a time-dependent manner (Fig. 5). Consistently, the steady-state Hmgcr mRNA level in hepatocytes was not diminished upon miR-27a overexpression (Fig. 5). However, Hmgcr mRNA levels were significantly altered at time points exceeding 24 h of transfection in hepatocytes or miR-27a mimic injection in Apoe−/− mice (Fig. 4B and Fig. 9C). These observations are consistent with a previous report that suggests that miRNAs initially reduce target protein levels without affecting mRNA levels but diminish mRNA levels at later time points (30). Therefore, we speculate that miR-27a-mediated Hmgcr repression mainly involves translational control followed by mRNA degradation.
In view of the key role of miR-27a in regulating Hmgcr, we sought to unravel how miR-27a might be regulated under basal and pathophysiological conditions. Computational and experimental analyses suggested a crucial role of Egr1 in the activation of miR-27a expression. Egr1, a zinc finger transcription factor belonging to the early growth response gene family, binds to a GC-rich consensus region (31) and regulates genes involved in the physiological stress response, cell metabolism, proliferation, and inflammation (32). Egr1, predominantly expressed in the liver, targets multiple cholesterol biosynthesis genes (11, 33). In light of the putative Egr1 binding sites in the proximal miR-27a promoter domain (Fig. 7A), we investigated the role of Egr1 in miR-27a expression. Indeed, the overexpression or downregulation of Egr1 resulted in enhanced or diminished miR-27a promoter activity, respectively, in AML12 cells (Fig. 7D and E). ChIP assays also confirmed the in vivo interaction of Egr1 with the miR-27a promoter in the context of chromatin (Fig. 7F). Furthermore, our results suggest that Egr1 regulates miR-27a expression under elevated cholesterol conditions (Fig. 7J).
Role of miR-27a in global regulation of cholesterol homeostasis.
miR-27a, an intergenic miRNA, is transcribed from the miR-23a–miR-27a–miR-24 cluster located on chromosome 8 in mice. Dysregulation of miR-27a has been associated with several cardiovascular phenotypes, including impaired left ventricular contractility, hypertrophic cardiomyopathy, adipose hypertrophy, and hyperplasia (34). miR-27a has been reported to regulate several genes involved in adipogenesis and lipid metabolism, including retinoid X receptor alpha (RXRα); ATP binding cassette transporter (ABCA1), also known as cholesterol efflux regulatory protein; fatty acid synthase (FASN); sterol regulatory element binding proteins (SREBP-1 and -2); peroxisome proliferator-activated receptors (PPAR-α and -γ), apolipoprotein A-1, apolipoprotein B-100, and apolipoprotein E-3 (35). In addition, miR-27a has been reported to play a role in the regulation of Ldlr (36). Our in vitro and in vivo experiments revealed that miR-27a targets Mvk, Fdft1, Sqle, Ggps1, and Mvd in the cholesterol biosynthesis pathway (Fig. 9). Consistent with our findings, administration of the miR-27a mimic in Apoe−/− mice diminished lipid levels in both plasma and peritoneal macrophages, thereby alleviating atherosclerosis by targeting macrophage-derived lipoprotein lipase (Lpl) (37). Of note, HITS-CLIP experiments revealed that miR-27a also targeted microsomal triglyceride transfer protein (Mttp), a crucial chaperone involved in lipoprotein production (38). Furthermore, global pathway analysis for miR-27a targets revealed 21 pathways, including steroid biosynthesis, fatty acid metabolism, and biosynthesis (Fig. 9D). Thus, miR-27a contributes to the global regulation of cholesterol homeostasis by targeting multiple genes in lipid synthesis, lipoprotein synthesis, and uptake.
Limitations of the study.
Although our findings indicate that the miR-27a mimic lowers plasma cholesterol levels considerably, there are some challenges that remain unaddressed. First, a less invasive or noninvasive method of delivery (viz., the subcutaneous or oral route) rather than intravenous injections would be desirable for easy administration of the mimic. Second, improved lipid formulations or nanoparticle-mediated delivery would considerably reduce the amount of mimic required and eliminate the need for frequent injections. Additionally, further chemical modifications to this mimic may increase target specificity and confer resistance to nuclease activity. Moreover, extensive assessments of the biological and pharmacological effects of the miR-27a mimic in animal models are necessary to evaluate its long-term safety and efficacy as a therapeutic agent.
Conclusions and perspectives.
This study identified miR-27a as a crucial regulator of the cholesterol biosynthesis pathway. Egr1 modulates miR-27a expression under basal and elevated-cholesterol conditions. miR-27a directly interacts with the 3′-UTR of Hmgcr and represses Hmgcr protein level by translational attenuation followed by mRNA decay. miR-27a augmentation in high-cholesterol-diet-fed Apoe−/− mice diminished plasma lipid and hepatic Hmgcr levels. These findings provide novel insights into the plausible role of miR-27a in the posttranscriptional regulation of Hmgcr, thereby implicating its role in cholesterol homeostasis under pathophysiological conditions.
miRNA therapeutics is an emerging and promising avenue to treat a wide array of human diseases. A fine balance between lipid/lipoprotein synthesis and lipoprotein uptake is essential for cholesterol homeostasis, and dysregulation of one or more pathways could lead to drastic effects. Therefore, miRNAs targeting multiple related pathways may be more efficacious in lowering plasma lipid levels than the conventional approach of targeting individual proteins or pathways. In view of the ability of miR-27a to target several crucial genes in the cholesterol biosynthesis pathway, the miR-27a mimic may emerge as a promising therapeutic intervention to lower plasma cholesterol levels. Thus, our findings and those of other studies provide a strong impetus for further evaluation of miR-27a as a novel lipid-lowering agent.
MATERIALS AND METHODS
Comparative genomics analyses.
For rat QTL analysis, data pertaining to elevated lipid/cholesterol QTLs, their respective LOD scores, and all the genes with their respective positions in a particular QTL were mined from the Rat Genome Database (Table 1). Each QTL is assigned a “logarithm of odds” (LOD) score, which is a measure of the strength of the evidence for the presence of a QTL at a particular location and its association with a trait. The elevated lipid/cholesterol QTLs in rats were plotted against the respective LOD scores. Comparative genome analysis of the rat genomic regions with mouse and human genomic regions at the Hmgcr locus was performed using mVISTA. The mVISTA browser uses the AVID alignment algorithm, which employs global alignment for sequence comparison of the input queries.
In silico predictions of potential miRNA binding sites in the Hmgcr 3′ UTR and putative miRNA targets in the cholesterol biosynthesis pathway.
Putative miRNA binding sites in the mouse Hmgcr (Hmgcr) 3′-UTR sequence (NCBI RefSeq accession number NM_008255.2) were predicted using various bioinformatic algorithms (viz., miRWalk, miRanda, TargetScan, PITA, RNA22, and RNAhybrid [Table 3]). Since a large number of miRNAs were predicted by these online tools, we selected only those miRNAs that were predicted by at least five algorithms. Furthermore, differences in hybridization free energy, indicating the stability of the microRNA-mRNA interaction, were determined computationally by two online tools called PITA and RNAhybrid. The lower or more negative ΔΔG value predicted by PITA indicates stronger binding of the microRNA to a given site; as a rule of thumb, sites with ΔΔG values below −10 are likely to be functional and were selected for experimental validation. RNAhybrid calculates the minimum free energy (ΔG) of hybridization between the target mRNA and miRNA. An RNAhybrid ΔG score of less than −20 kcal/mol is a strong indicator of interactions of an miRNA with the target mRNA. In each case, the minimum number of nucleotides in the seed sequence was selected as 6, and no mismatch or G-U wobble base pairing was allowed in the region of the seed sequence. The lower or more negative ΔG (less than −10) or ΔΔG (less than −20) values predicted by PITA and RNAhybrid, in general, indicate stronger binding of the miRNA to the target gene 3′ UTR.
In order to predict miR-27a target genes in the cholesterol biosynthesis pathway, putative miR-27a targets were retrieved from TargetScan and miRWalk (Table 3). These targets were grouped by the PANTHER classification system (http://www.pantherdb.org/) based on their molecular functions, and the genes mapping to the cholesterol biosynthesis pathway were selected. In addition, genes involved in the cholesterol biosynthesis pathway having an RNAhybrid ΔΔG score of less than −20 were also considered putative miR-27a targets.
Tissue-specific expression of endogenous HMGCR, hsa-miR-27a-3p, hsa-miR-28, and hsa-miR-708 in human tissues.
For correlation analysis, human tissues that showed consistent HMGCR expression across the GTEx, BioGPS, and SAGE (Table 3) databases were selected for analysis. The expression of HMGCR across various human tissues was mined from the GTEx portal (Table 3). Likewise, tissue-specific hsa-miR-27a-3p expression was obtained from DASHR (Table 3). Furthermore, tissues showing hsa-miR-27a-3p expression in the range of 100 to 1,000 RPM were selected since miRNAs expressed in this range were reported to bring about significant target repression (10). Only tissues common to the GTEx portal and DASHR were chosen for correlation analysis. The data were normalized to a particular tissue (having the lowest expression level of hsa-miR-27a-3p) and expressed as fold changes. Specifically, the expression data from DASHR and GTEx were normalized to spleen values. Likewise, the tissues that were used for correlation analysis of hsa-miR-27a-3p and HMGCR expression were also analyzed for hsa-miR-28 and hsa-miR-708 expression.
Pathway analysis for miR-27a targets.
In order to identify pathways that may be targeted by miR-27a, the TargetScan and mirPath v3 tools were employed (Table 3). TargetScan relies on an in silico prediction of miRNA targets in a specific pathway, while mirPath v3 employs prediction algorithms and experimentally validated miRNA-gene interactions. Interestingly, 21 pathways that were predicted miR-27a targets were commonly enriched according to both these tools, including steroid biosynthesis, fatty acid metabolism, and biosynthesis.
Generation of Hmgcr 3′-UTR–luciferase, mmu-miR-27a promoter–luciferase reporter constructs, and miRNA expression plasmids.
To generate the Hmgcr 3′-UTR–luciferase construct, the mouse Hmgcr 3′-UTR domain (bp +20359 to +21975 bp) was PCR amplified using Phusion high-fidelity DNA polymerase (Finnzymes), mouse genomic DNA (Jackson Laboratory, Bar Harbor, ME, USA), and gene-specific primers (forward primer [FP], 5′-CGTGCTAGCGGATCCTGACACTGAACTG-3′; reverse primer [RP], 5′-GCGGCCGGCCTTCAATGTTAACTTCCTTTC-3′). The numberings of the nucleotide positions are with respect to the cap site as the +1 position. Underlined nucleotides in the forward and reverse primer sequences are the restriction sites for NheI and FseI, respectively, which were added to enable the cloning of the PCR-amplified 3′ UTR into the firefly luciferase-expressing pGL3-promoter reporter vector (Promega). The purified Hmgcr 3′-UTR PCR product was cloned between the XbaI and FseI sites of the pGL3-promoter vector because the NheI-digested PCR product had compatible ends for the XbaI-digested pGL3-promoter reporter vector. The authenticity of the Hmgcr 3′-UTR reporter plasmid was confirmed by DNA sequencing using pGL3-promoter vector sequencing primers (forward primer, 5′-CGTCGCCAGTCAAGTAACAA-3′ [bp 1782 to 1801]; and reverse primer, 5′-CCCCCTGAACCTGAAACATA-3′ [bp 2118 to 2137]); the resultant plasmid was named the mHmgcr 3′ UTR plasmid.
To abrogate the binding of miR-27a, a 3′-UTR deletion construct was generated using site-directed mutagenesis wherein the putative miR-27a binding site was deleted. The 3′-UTR deletion construct for miR-27a was generated by using the wild-type Hmgcr-3′-UTR reporter construct as the template and the following primers: forward primer 5′-CGCGGGCATTGGGTTCTCAATTAAAAATCTCAATGCACT-3′ and reverse primer 5′-AGTGCATTGAGATTTTTAATTGAGAACCCAATGCCCGCG-3′. This deletion in the reporter plasmid was confirmed by DNA sequencing, and the resultant construct was named mHmgcr 27a mut 3′UTR.
The mmu-miR-27a promoter–luciferase reporter construct was generated by amplifying the region spanning bp −1079 to +26 of the miR-27a promoter using mouse genomic DNA as described above and primers (forward primer, 5′-CTAGCTAGCAACTTTAACTGGCACGCAGG-3′; reverse primer, 5′-CCGCTCGAGGGCATCAAATCCCATCCC-3′). Underlined nucleotides in the forward and reverse primers are the restriction sites for NheI and XhoI, respectively, which were added to assist in the cloning of the PCR-amplified miR-27a promoter region into the pGL3-basic vector (Promega). The authenticity of the resultant construct (referred to as the miR-27a promoter) was confirmed by DNA sequencing.
To generate a plasmid expressing miR-27a, the sequence of the pre-miRNA was retrieved from the miR-Base/UCSC genome browser, PCR amplified using mouse genomic DNA as the template, and primers (forward primer, 5′-CGCGGATCCTCGCCAAGGATGTCTGTCTT-3′; reverse primer, 5′-CCGCTCGAGGTTTCAGCTCAGTAGGCACG-3′). Underlined nucleotides are the BamHI and XhoI restriction sites that were added to the forward and reverse primers, respectively. The purified PCR-amplified DNA was cloned between the BamHI and XhoI restriction sites in the pcDNA3.1 vector (Invitrogen). The resultant plasmid was named the miR-27a expression plasmid, and the accuracy of the insert was confirmed by DNA sequencing using miRNA-specific primers. Similarly, miR-27b and miR-764 expression plasmids were generated using specific primers (forward primer 5′-CGCGGATCCTGCAGTTTGGAGAACAGAGG-3′ and reverse primer 5′-CCGCTCGAGCTTGAGGCAGGCTGGTCT-3′, and forward primer 5′-CGCGGATCCCCTTGTGGTATTGTTGGAAGG-3′ and reverse primer 5′-CCGCTCGAGTCTTTCCTTTGCTCTACCTTG-3′, respectively), harboring BamHI and XhoI restriction sites to enable cloning into the pcDNA3.1 vector (Invitrogen).
Cell lines, transfections, and reporter assays.
AML12 (alpha mouse liver 12) (a gift from Rakesh K. Tyagi, Jawaharlal Nehru University, New Delhi, India), mouse neuroblastoma N2a, and human hepatocellular carcinoma HuH-7 (obtained from the National Center for Cell Sciences, Pune, India) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with high glucose and glutamine (HyClone), supplemented with 10% fetal bovine serum (Invitrogen, USA), penicillin G (100 U/ml), and streptomycin sulfate (100 mg/ml) (Invitrogen), in 25-cm2 tissue culture flasks (Nest) at 37°C with 5% CO2. These cell lines were tested for mycoplasma infection routinely and treated with BM-cyclin (Merck) to eliminate mycoplasma, if detected.
AML12 and HuH-7 cells were grown to 70% confluence in 12-well plates and cotransfected with different doses (125, 250, and 500 ng) of the miR-27a expression plasmid, along with 500 ng/well of the mHmgcr 3′-UTR plasmid by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Similarly, the mHmgcr 3′-UTR plasmid or the mHmgcr 27a mut 3′-UTR construct was cotransfected with the miR-27a expression plasmid in a dose-dependent manner. In all these cotransfection experiments, the insert-free vector pcDNA3.1 was used as a balancing plasmid. After 4 h of transfection, the culture medium was changed to fresh complete medium.
In other cotransfection experiments, AML12 cells were transfected with different doses of an Egr1 expression plasmid (obtained from Donna Lee Wong) (39) or Egr1 short hairpin RNA (shRNA) expression plasmids (obtained from Weihua Xiao) (40), along with 500 ng/well of the miR-27a promoter construct. In order to maintain equal amounts of DNA across transfections, pcDNA3.1 and pU6 were used as balancing plasmids with the Egr1 and Egr1 shRNA expression plasmids, respectively.
In all cotransfection experiments, cells were lysed at 36 h posttransfection, and cell lysates were assayed for luciferase activity. Luciferase assays were carried out as described previously (41, 42). The total amount of protein per individual well was also estimated in the same cell lysate using Bradford reagent (Bio-Rad). The reporter activities were normalized to the total protein content and expressed as luciferase activity per microgram of protein or a percentage over the control.
Animals and tissue samples.
All animal-related procedures were approved by the Institutional Animal Ethics Committee of the Indian Institute of Technology Madras, Chennai, India, as well as CSIR-Central Drug Research Institute, Lucknow, India, and performed in accordance with the Guide for the Care and Use of Laboratory Animals, 8th edition, of the U.S. National Institutes of Health (43) and ARRIVE (Animals in Research: Reporting In Vivo Experiments) guidelines (44). Male Apoe−/− mice (18 to 20 g and 8 to 10 weeks old) on a C57BL/6 background were obtained from the National Laboratory Animal Centre, CSIR-Central Drug Research Institute, Lucknow, India. The animals were maintained in polypropylene cages under controlled room temperature at 25°C on a 12-h light-dark cycle. The mice were fed a high-cholesterol diet (HCD) containing 0.21%, 20%, 50%, and 21% cholesterol, protein, carbohydrate, and fat (by weight), respectively (Research Diets) for 10 weeks. All mice received water and food ad libitum. After completion of the 10-week diet regimen, the mice were randomly assigned to three groups: HCD-fed mice injected with either saline (n = 7), the miRVANA miR-27a mimic (Invitrogen) (n = 7), or miRVANA negative-control oligonucleotides (Invitrogen) (n = 7).
The miR-27a mimic and negative-control oligonucleotides were synthesized by Invitrogen, complexed with Invivofectamine 3.0 (Invitrogen), and injected into the tail vein at a dose of 5 mg/kg body weight twice over a period of 11 days (with the second injection a week after the first dosing). Invivofectamine 3.0 is a lipid nanoparticle delivery system used for the in vivo delivery of miRNAs or small interfering RNAs (siRNAs) in rodents. Owing to its safety and efficacy, multiple studies have reported the use of Invivofectamine 3.0 for the delivery of miRNA mimics or inhibitors in vivo in mice (25, 45–47). For the in vivo delivery of the miR-27a mimic or control oligonucleotide, 75 μl of a 19.2-μg/μl solution of the control oligonucleotide or miR-27a mimic was added to an equal volume of complexation buffer. This mixture was then diluted with an equal volume of Invivofectamine 3.0 and vortexed for a few seconds to ensure miR-27a–Invivofectamine complex formation. The complexes were incubated at 50°C for 30 min. These complexes were then diluted appropriately with phosphate-buffered saline (PBS) to attain a final concentration of 0.5 mg/ml. Approximately 200 μl of this formulation was injected into Apoe−/− mice via the tail vein. The injected mice were still continued on an HCD until they were sacrificed. The body weight of the animals was monitored every alternate day. Four days after the second dosing, the animals were euthanized by CO2 inhalation followed by cardiac puncture (for blood collection into heparin vials), and the organs were collected in RNAlater (Thermo Fisher) and neutral buffered saline and stored at −80°C for further analyses. Wistar female rats at the age of 6 weeks were obtained from the King Institute of Preventive Medicine (Chennai, India). Liver, kidneys, heart, and skeletal muscle were isolated according to standard procedures.
Analysis of biochemical parameters in plasma, blood, and tissue samples from miRNA- or control oligonucleotide-treated mice.
Animals were fasted overnight, and plasma was collected by spinning whole blood at 2,000 rpm at 4°C for 20 min to measure total, HDL, and LDL cholesterol levels using commercial kits (Randox) according to the manufacturer’s instructions. Plasma alanine aminotransferase (ALT), aspartate aminotransferase (AST), and creatine kinase (CK) levels were also measured using kits (Randox) according to the manufacturer’s protocol. For estimating hepatic cholesterol levels, liver tissue (∼20 mg) was homogenized in a solvent containing hexane-isopropanol at a ratio of 3:2. The lipids were then extracted, and tissue total cholesterol was assayed by using an Amplex red cholesterol assay kit (Thermo Fisher) according to the manufacturer’s protocol. For fasting blood glucose level measurements, blood was collected via retro-orbital bleed after fasting the animals overnight, and the blood glucose levels were measured using an Accu-Chek active blood glucometer (Roche).
RNA extraction and real-time PCR.
Total RNA was extracted from cell lines and tissue samples by using TRIzol (Invitrogen) according to the manufacturer’s instructions. cDNA synthesis was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems) and miR-27a-, miR-27b-, or U6-specific stem-loop (SL) (Table 4) or random hexamer primers. Quantitative real-time PCR (qPCR) was carried out using the DyNAmo HS-SYBR green qPCR kit (Finnzymes), miR-27a/b primers, and a universal reverse primer or gene-specific primers (Table 4). The same mmu-miR-27 forward primer was also used to probe for miR-27b expression.
TABLE 4.
Primers used for qPCR analysesa
| Primer | Sequence (5′–3′) |
|---|---|
| miR-27a-SL-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCGGAAC |
| miR-27b-SL-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCAGAAC |
| U6-SL-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTATGGAAC |
| mmu-miR-27-FP | ACACTTTCACAGTGGCTAA |
| U6-FP | CTGCGCAAGGATGACACGCA |
| Universal RP | GTGCAGGGTCCGAGGT |
| mHmgcr-FP | GAGAATGCAGAGAAAGGTG |
| mHmgcr-RP | GGCGAATAGACACACCAC |
| mβ-Actin-FP | CTTCTTTGCAGCTCCTTCGTT |
| mβ-Actin-RP | TTCTGACCCATTCCCACCA |
| rHmgcr-FP | ACCTGCTGCCATAAACTGGAT |
| rHmgcr-RP | ACCACCTTGGCTGGAATGAC |
| rβ-Actin-FP | GCTGTGCTATGTTGCCCTAG |
| rβ-Actin-RP | CGCTCATTGCCGATAGTG |
| hHmgcr-FP | TCGGTGGCCTCTAGTGAGAT |
| hHmgcr-RP | TGTCCCCACTATGACTTCCC |
| mMvk-FP | GGTGTGGTCGGAACTTCCC |
| mMvk-RP | CCTTGAGCGGGTTGGAGAC |
| mFdft1-FP | ATGGAGTTCGTCAAGTGTCTAGG |
| mFdft1-RP | CGTGCCGTATGTCCCCATC |
| mHmgcs1-FP | TTGAGGAGTCTGGGAATACAG |
| mHmgcs1-RP | CATATCGTCCATCCCAAGAG |
| mGgps1-FP | GTCATCTCCAGCAGTTCCTTC |
| mGgps1-RP | TCATCTCGTCCAGCATCTTC |
| mScarb1-FP | CAGGTGCTCAAGAATGTCC |
| mScarb1-RP | TTTGTCTGAACTCCCTGTAG |
| mMvd-FP | ATGGCCTCAGAAAAGCCTCAG |
| mMvd-RP | TGGTCGTTTTTAGCTGGTCCT |
Abbreviations: FP, forward primer; RP, reverse primer; SL, stem-loop.
In another set of experiments, AML12 cells were transfected with different doses of the miR-27a expression plasmid or 1 μg of the miRVANA miR-27a mimic (Invitrogen) and miRVANA negative-control oligonucleotides (Invitrogen) or with 60 nM locked nucleic acid inhibitor of 27a (LNA 27a) and negative-control oligonucleotides (Exiqon) using Lipofectamine. The overexpression or downregulation of miR-27a was determined by qPCR analysis.
For cholesterol depletion, AML12 cells (grown in 12-well plates) were treated with increasing doses (0, 1, 2.5, and 5 mM) of the cholesterol-depleting reagent methyl-β-cyclodextrin (MCD) (HiMedia) for 15 min. Cholesterol depletion was carried out in serum-free DMEM. Following cholesterol depletion, the medium was changed to fresh serum-free medium, and the cells were incubated for 6 to 9 h at 37°C in a CO2 incubator. In another series of experiments, AML12 cells were treated with exogenous cholesterol (0, 5, 10, and 20 μg/ml) in serum-free medium for 6 to 9 h. Next, the cells were processed for RNA isolation followed by qPCR to measure the relative abundances of miR-27a and Hmgcr transcripts. Total miRNAs isolated from the treated and control cells were subjected to cDNA synthesis followed by qPCR analysis probing for miR-27a and U6 RNA using miR-27a- and U6-specific primers. In all the qPCR analyses, the relative abundances of miR-27a and Hmgcr were determined by calculating the value for each reaction (48).
Filipin staining of hepatocyte cells.
AML12 cells were seeded at 60 to 70% confluence in 12- or 24-well plates overnight at 37°C with 5% CO2. The cells were then treated with 20 μg/ml of exogenous cholesterol or 5 mM the cholesterol-depleting reagent MCD for 6 h or 15 min, respectively. After MCD treatment, the medium was changed to serum-free medium, and the cells were incubated at 37°C for 6 to 9 h. Following treatment with cholesterol or MCD, the cells were washed with PBS and fixed with 3.6% formaldehyde in PBS for 10 min at room temperature. The fixed cells were washed with PBS and stained with 50 μg/ml of filipin III (a fluorescent dye that binds to free cholesterol) in the dark at room temperature for 2 h. Following fixation, the cells were washed with PBS again and imaged using an Olympus U-RFL-T fluorescence microscope.
mRNA stability assays.
Actinomycin D, an extensively used and highly specific transcriptional inhibitor, was used to determine Hmgcr mRNA stability as described previously (49). In this experiment, AML12 cells were transfected with the miR-27a plasmid or pcDNA3.1. After 12 h of transfection, cells were treated with actinomycin D (5 μg/ml) (HiMedia) for different times (0, 6, 12, and 24 h). In both cases (with and without miR-27a overexpression), Hmgcr mRNA decay was monitored by measuring the Hmgcr levels by qPCR. The Hmgcr mRNA half-life was determined by using the formula t1/2 = −t(ln2)/ln(Nt/N0), where Nt is the mRNA remaining at a specific time, t, and N0 is the mRNA abundance at the beginning of the experiment. Western blot analysis for Hmgcr levels at each time point following actinomycin D treatment was also performed.
Pulse-chase analysis and immunoprecipitation assays.
Pulse-chase analysis was performed as previously described (50). In brief, HuH-7 cells were transfected with either the miR-27a expression plasmid or pcDNA3.1 in 35-mm cell culture dishes. The cells were starved for an hour, followed by labeling with [35S]Met (BRIT, Mumbai, India) for 1 h (25 μCi/35-mm dish). The cells were chased for 30, 60, and 120 min, and cell lysates were immunoprecipitated using the HMGCR antibody (catalog number ab174830; Abcam). The immunoprecipitated lysates were resolved by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Pall Life Sciences) and analyzed by autoradiography.
Ago2-ribonucleoprotein immunoprecipitation assays.
Ago2-ribonucleoprotein immunoprecipitation (RIP) assays were performed as described previously (51). HuH-7 cells grown to 60% confluence in 100-mm dishes were transfected with 5 μg of the miR-27a plasmid or pcDNA3.1 using Targetfect F2 transfection reagent (Targeting Systems). After 24 h of transfection, cells were lysed in 100 μl of ice-cold polysome lysis buffer (5 mM MgCl2, 100 mM KCl, 10 mM HEPES [pH 7.0], and 0.5% Nonidet P-40) with freshly added 1 mM dithiothreitol (DTT) and 100 U/ml recombinant RNase inhibitor (TaKaRa) and supplemented with a protease inhibitor cocktail (Sigma-Aldrich) by tapping every 5 min for 3 s over a period of 15 min on ice. The lysates were then centrifuged at 14,000 rpm at 4°C for 10 min. The supernatant was mixed with 900 μl of ice-cold NT2 buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40) containing freshly added 200 U/ml recombinant RNase inhibitor (TaKaRa), 1 mM DTT, and 15 mM EDTA. The lysates were precleared with Rec protein G-Sepharose 4B beads (Invitrogen). Precleared samples were then immunoprecipitated by incubation overnight at 4°C with either 0.75 μg of anti-Ago2 antibody (catalog number ab57113; Abcam) or nonimmune mouse IgG (catalog number I5831; Sigma). On the following day, beads were washed five times with ice-cold NT2 buffer and divided into two fractions: one for RNA isolation to identify miRNA target genes and another for Western blotting to check for successful immunoprecipitation of Ago2. Anti-Ago2 antibody at a dilution of 1:2,500 was used for Western blot analysis. RNA was isolated from the other fraction of the beads by using TRIzol (Invitrogen), followed by purification via NucleoSpin miRNA columns (Macherey-Nagel).
Western blot analysis.
After transfection, cholesterol treatment, or depletion experiments, AML12 or HuH-7 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.2], 150 mM NaCl, 1% [vol/vol] Triton X-100, 1% [wt/vol] sodium deoxycholate, 1 mM EDTA, and 0.1% [wt/vol] SDS) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and a protease inhibitor cocktail (Sigma). The cell lysates were sonicated for 10 to 15 s on ice, followed by centrifugation at 14,000 rpm for 15 min at 4°C, and the supernatant was then collected. For tissue protein isolation, the liver tissue samples were washed with PBS and homogenized in 1.0 ml RIPA buffer using a micropestle (Tarsons) in a 1.5-ml microcentrifuge tube. The homogenized samples were sonicated and centrifuged at 14,000 rpm, and the supernatant was stored in aliquots at −80°C until further use. The protein concentrations in the cell lysates or tissues were estimated by a Bradford assay (Bio-Rad). Equal amounts of protein samples (∼30 to 50 μg) under each condition were separated in an SDS-PAGE gel and transferred to an activated PVDF membrane (Pall Life Sciences). After blocking with 2 to 5% bovine serum albumin (BSA) or nonfat milk for 1 h at room temperature, the membranes were incubated with a specific primary antibody (for HMGCR [catalog number ab174830; Abcam] at a 1:1,000 dilution, β-actin [catalog number A5441; Sigma] at a 1:7,500 dilution, vinculin [catalog number V9131; Sigma] at a 1:7,500 dilution, or Egr1 [catalog number 4153; CST] at a 1:1,000 dilution) overnight at 4°C. After washing with 1× Tris-buffered saline–Tween (TBST), the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibody specific for either rabbit (catalog number 170-6515; Bio-Rad) (at a 1:1,500 dilution for HMGCR or Egr1) or mouse (catalog number 115-035-003; Jackson Immunoresearch) (at a 1:5,000 dilution for β-actin) for 1 h. The protein bands were detected using a Clarity Western ECL substrate kit (Bio-Rad), and the signal was captured by using the Chemidoc XRS+ chemiluminescence detection system (Bio-Rad). Densitometric analysis of Western blots was performed using Image Lab (Bio-Rad) or NIH ImageJ software (52). All the Western blot experiments were repeated at least three times, and representative images are shown.
Chromatin immunoprecipitation assays.
AML12 cells at 60 to 80% confluence were cross-linked using formaldehyde at room temperature for 12 min. Next, chromatin was isolated and sheared by sonication, followed by a preclearing step with Rec protein G-Sepharose 4B beads (Invitrogen). Immunoprecipitation reactions with the precleared samples were carried out by incubation with 5 μg each of chromatin immunoprecipitation (ChIP)-grade antibodies, i.e., anti-Egr1 and preimmune anti-rabbit IgG (catalog number I5006; Sigma), overnight at 4°C. The immunoprecipitated samples were captured by Rec protein G-Sepharose 4B beads, eluted, reverse cross-linked, and purified by phenol-chloroform extraction. qPCR was carried out using an equal amount of the purified chromatin as the template to amplify two different DNA regions harboring Egr1 binding sites in the proximal (∼500-bp) promoter domain of the miR-27a promoter using two primer pairs (Fig. 7A) (P1-FP [5′-TCAAGATAGGCAGGCAAGC-3′] and P1-RP [5′-AGCACAGGGTCAGTTGGAAA-3′], and P2-FP [5′-TTTGTAGGGCTGGGGTAGAG-3′] and P2-RP [5′-CTGATCCACACCCTAGCCC-3′]). Results are expressed as fold enrichment over the IgG signal or background.
Data presentation and statistical analysis.
All transient-transfection experiments were performed at least three times, and results are expressed as means ± standard errors of the means (SEM) from triplicates. The Prism 5 program (GraphPad Software, USA) was used to determine the level of statistical significance by Student’s t test or one-way analysis of variance (ANOVA) with a Newman-Keuls posttest, as appropriate.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the Council of Scientific and Industrial Research (CSIR), Government of India, to N.R.M. [project number 37(1564)/12-EMR-II]. This work was also partly supported by an exploratory research project grant from Industrial Consultancy and Sponsored Research, IIT Madras. Research fellowships were received from the Ministry of Human Resource Development (to A.A.K. and V.G.), the Department of Science and Technology (to V.A.), the Indian Council of Medical Research (to S.S.R. and H.A.), and the CSIR (to A.K. and B.N.), Government of India.
We are grateful to Dona Lee Wong (Harvard Medical School, Boston, MA) for providing the Egr1 expression plasmid and to Weihua Xiao (University of Science and Technology of China, Hefei, China) for the Egr1 shRNA plasmid. We also thank Rakesh K. Tyagi, Jawaharlal Nehru University, New Delhi, India, for providing the AML12 cell line and Madhu Dikshit, Translational Health Science and Technology Institute, Faridabad, India, for help at the initial phase of this study. We also thank V. Janani (IIT Madras, India) for her technical support with the manuscript.
We declare that we have no conflict of interest.
REFERENCES
- 1.Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, de Ferranti SD, Ferguson JF, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu SM, Longenecker CT, Lutsey PL, Mackey JS, Matchar DB, Matsushita K, Mussolino ME, Nasir K, O’Flaherty M, Palaniappan LP, Pandey A, Pandey DK, Reeves MJ, Ritchey MD, Rodriguez CJ, Roth GA, Rosamond WD, Sampson UKA, Satou GM, Shah SH, Spartano NL, Tirschwell DL, Tsao CW, Voeks JH, Willey JZ, Wilkins JT, Wu JHY, Alger HM, et al. . 2018. Heart disease and stroke statistics—2018 update: a report from the American Heart Association. Circulation 137:e67–e492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 2.Sathiyakumar V, Kapoor K, Jones SR, Banach M, Martin SS, Toth PP. 2018. Novel therapeutic targets for managing dyslipidemia. Trends Pharmacol Sci 39:733–747. doi: 10.1016/j.tips.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Athyros VG, Doumas M, Imprialos KP, Stavropoulos K, Georgianou E, Katsimardou A, Karagiannis A. 2018. Diabetes and lipid metabolism. Hormones (Athens) 17:61–67. doi: 10.1007/s42000-018-0014-8. [DOI] [PubMed] [Google Scholar]
- 4.Palacio Rojas M, Prieto C, Bermúdez V, Garicano C, Núñez Nava T, Martínez MS, Salazar J, Rojas E, Pérez A, Marca Vicuña P, González Martínez N, Maldonado Parra S, Hoedebecke K, D’Addosio R, Cano C, Rojas J. 2017. Dyslipidemia: genetics, lipoprotein lipase and HindIII polymorphism. F1000Res 6:2073. doi: 10.12688/f1000research.12938.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geelen MJH, Gibson DM, Rodwell VW. 1986. Hydroxymethylglutaryl-CoA reductase—the rate-limiting enzyme of cholesterol biosynthesis. A report of a meeting held at Nijenrode Castle, Breukelen, The Netherlands, August 24, 1985. FEBS Lett 201:183–186. doi: 10.1016/0014-5793(86)80604-4. [DOI] [PubMed] [Google Scholar]
- 6.Stone NJ, Robinson JG, Lichtenstein AH, Merz CNB, Blum CB, Eckel RH, Goldberg AC, Gordon D, Levy D, Lloyd-Jones DM, McBride P, Schwartz JS, Shero ST, Smith SC Jr, Watson K, Wilson PWF, American College of Cardiology/American Heart Association Task Force on Practice Guidelines . 2014. 2013 ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol 63:2889–2934. doi: 10.1016/j.jacc.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 7.DeBose-Boyd RA. 2008. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res 18:609–621. doi: 10.1038/cr.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemecz M, Alexandru N, Tanko G, Georgescu A. 2016. Role of microRNA in endothelial dysfunction and hypertension. Curr Hypertens Rep 18:87. doi: 10.1007/s11906-016-0696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condorelli G, Latronico MVG, Cavarretta E. 2014. MicroRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol 63:2177–2187. doi: 10.1016/j.jacc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 10.Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. 2012. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods 9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gokey NG, Lopez-Anido C, Gillian-Daniel AL, Svaren J. 2011. Early growth response 1 (Egr1) regulates cholesterol biosynthetic gene expression. J Biol Chem 286:29501–29510. doi: 10.1074/jbc.M111.263509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang M, Liu W, Pellicane C, Sahyoun C, Joseph BK, Gallo-Ebert C, Donigan M, Pandya D, Giordano C, Bata A, Nickels JT Jr. 2014. Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J Lipid Res 55:226–238. doi: 10.1194/jlr.M041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramírez CM, Rotllan N, Vlassov AV, Dávalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, Wanschel A, Zavadil J, Castrillo A, Kim J, Suárez Y, Fernández-Hernando C. 2013. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ Res 112:1592–1601. doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurtz CL, Fannin EE, Toth CL, Pearson DS, Vickers KC, Sethupathy P. 2015. Inhibition of miR-29 has a significant lipid-lowering benefit through suppression of lipogenic programs in liver. Sci Rep 5:12911. doi: 10.1038/srep12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia YM, Ling MF, Zhang LC, Jiang SX, Sha YS, Zhao RQ. 2016. Downregulation of miR-150 expression by DNA hypermethylation is associated with high 2-hydroxy-(4-methylthio)butanoic acid-induced hepatic cholesterol accumulation in nursery piglets. J Agric Food Chem 64:7530–7539. doi: 10.1021/acs.jafc.6b03615. [DOI] [PubMed] [Google Scholar]
- 16.Zhou L, Hussain MM. 2017. Human microRNA-548p decreases hepatic apolipoprotein B secretion and lipid synthesis. Arterioscler Thromb Vasc Biol 37:786–793. doi: 10.1161/ATVBAHA.117.309247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun C, Huang F, Liu X, Xiao X, Yang M, Hu G, Liu H, Liao L. 2015. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int J Mol Med 35:847–853. doi: 10.3892/ijmm.2015.2076. [DOI] [PubMed] [Google Scholar]
- 18.Singh R, Yadav V, Kumar S, Saini N. 2015. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci Rep 5:17454. doi: 10.1038/srep17454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selitsky SR, Dinh TA, Toth CL, Kurtz CL, Honda M, Struck BR, Kaneko S, Vickers KC, Lemon SM, Sethupathy P. 2015. Transcriptomic analysis of chronic hepatitis B and C and liver cancer reveals microRNA-mediated control of cholesterol synthesis programs. mBio 6:e01500-15. doi: 10.1128/mBio.01500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown MS, Goldstein JL. 2009. Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res 50:S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. 2002. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell 10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- 22.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP. 2012. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Zalzala M, Xu J, Li Y, Yin L, Zhang Y. 2015. miR-34a-HNF4α pathway regulates lipid and lipoprotein metabolism. Nat Commun 6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irani S, Iqbal J, Antoni WJ, Ijaz L, Hussain MM. 2018. MicroRNA-30c reduces plasma cholesterol in homozygous familial hypercholesterolemic and type 2 diabetic mouse models. J Lipid Res 59:144–154. doi: 10.1194/jlr.M081299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irani S, Pan X, Peck BC, Iqbal J, Sethupathy P, Hussain MM. 2016. MicroRNA-30c mimic mitigates hypercholesterolemia and atherosclerosis in mice. J Biol Chem 291:18397–18409. doi: 10.1074/jbc.M116.728451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang MY, Sun WL, Zhou MH, Tang Y. 2017. MicroRNA-27a regulates hepatic lipid metabolism and alleviates NAFLD via repressing FAS and SCD1. Sci Rep 7:14493. doi: 10.1038/s41598-017-15141-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shim J, Poulsen CB, Hagensen MK, Larsen T, Heegaard PMH, Christoffersen C, Bolund L, Schmidt M, Liu Y, Li J, Li R, Callesen H, Bentzon JF, Sorensen CB. 2017. Apolipoprotein E deficiency increases remnant lipoproteins and accelerates progressive atherosclerosis, but not xanthoma formation, in gene-modified minipigs. JACC Basic Transl Sci 2:591–600. doi: 10.1016/j.jacbts.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura T, Fabian MR. 2016. Scanning for a unified model for translational repression by microRNAs. EMBO J 35:1158–1159. doi: 10.15252/embj.201694324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 31.Christy B, Nathans D. 1989. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci U S A 86:8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu M, Zhu X, Zhang J, Liang J, Lin Y, Zhao L, Ehrengruber MU, Chen YE. 2003. Egr-1 target genes in human endothelial cells identified by microarray analysis. Gene 315:33–41. doi: 10.1016/s0378-1119(03)00730-3. [DOI] [PubMed] [Google Scholar]
- 33.Mohn KL, Laz TM, Melby AE, Taub R. 1990. Immediate-early gene-expression differs between regenerating liver, insulin-stimulated H-35 cells, and mitogen-stimulated Balb/C 3t3 cells. Liver-specific induction-patterns of gene-33, phosphoenolpyruvate carboxykinase, and the jun, fos, and egr families. J Biol Chem 265:21914–21921. [PubMed] [Google Scholar]
- 34.Devaux Y, Vausort M, McCann GP, Kelly D, Collignon O, Ng LL, Wagner DR, Squire IB. 2013. A panel of 4 microRNAs facilitates the prediction of left ventricular contractility after acute myocardial infarction. PLoS One 8:e70644. doi: 10.1371/journal.pone.0070644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, Cappello T, Wang L. 2015. Emerging role of microRNAs in lipid metabolism. Acta Pharm Sin B 5:145–150. doi: 10.1016/j.apsb.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez ML, Khosroheidari M, Eddy E, Done SC. 2015. MicroRNA-27a decreases the level and efficiency of the LDL receptor and contributes to the dysregulation of cholesterol homeostasis. Atherosclerosis 242:595–604. doi: 10.1016/j.atherosclerosis.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie W, Li L, Zhang M, Cheng HP, Gong D, Lv YC, Yao F, He PP, Ouyang XP, Lan G, Liu D, Zhao ZW, Tan YL, Zheng XL, Yin WD, Tang CK. 2016. MicroRNA-27 prevents atherosclerosis by suppressing lipoprotein lipase-induced lipid accumulation and inflammatory response in apolipoprotein E knockout mice. PLoS One 11:e0157085. doi: 10.1371/journal.pone.0157085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schug J, McKenna LB, Walton G, Hand N, Mukherjee S, Essuman K, Shi ZJ, Gao Y, Markley K, Nakagawa M, Kameswaran V, Vourekas A, Friedman JR, Kaestner KH, Greenbaum LE. 2013. Dynamic recruitment of microRNAs to their mRNA targets in the regenerating liver. BMC Genomics 14:264. doi: 10.1186/1471-2164-14-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta MP, Gupta M, Zak R, Sukhatme VP. 1991. Egr-1, a serum-inducible zinc finger protein, regulates transcription of the rat cardiac alpha-myosin heavy chain gene. J Biol Chem 266:12813–12816. [PubMed] [Google Scholar]
- 40.Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahapatra NR, Mahata M, Ghosh S, Gayen JR, O’Connor DT, Mahata SK. 2006. Molecular basis of neuroendocrine cell type-specific expression of the chromogranin B gene: crucial role of the transcription factors CREB, AP-2, Egr-1 and Sp1. J Neurochem 99:119–133. doi: 10.1111/j.1471-4159.2006.04128.x. [DOI] [PubMed] [Google Scholar]
- 42.Mahapatra NR, Mahata M, O’Connor DT, Mahata SK. 2003. Secretin activation of chromogranin A gene transcription. Identification of the signaling pathways in cis and in trans. J Biol Chem 278:19986–19994. doi: 10.1074/jbc.M207983200. [DOI] [PubMed] [Google Scholar]
- 43.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 44.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. 2010. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lesizza P, Prosdocimo G, Martinelli V, Sinagra G, Zacchigna S, Giacca M. 2017. Single-dose intracardiac injection of pro-regenerative microRNAs improves cardiac function after myocardial infarction. Circ Res 120:1298–1304. doi: 10.1161/CIRCRESAHA.116.309589. [DOI] [PubMed] [Google Scholar]
- 46.Schlosser K, Taha M, Stewart DJ. 2018. Systematic assessment of strategies for lung-targeted delivery of microRNA mimics. Theranostics 8:1213–1226. doi: 10.7150/thno.22912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmad N, Kushwaha P, Karvande A, Tripathi AK, Kothari P, Adhikary S, Khedgikar V, Mishra VK, Trivedi R. 2019. MicroRNA-672-5p identified during weaning reverses osteopenia and sarcopenia in ovariectomized mice. Mol Ther Nucleic Acids 14:536–549. doi: 10.1016/j.omtn.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski UH. 2008. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matassa DS, Amoroso MR, Agliarulo I, Maddalena F, Sisinni L, Paladino S, Romano S, Romano MF, Sagar V, Loreni F, Landriscina M, Esposito F. 2013. Translational control in the stress adaptive response of cancer cells: a novel role for the heat shock protein TRAP1. Cell Death Dis 4:e851. doi: 10.1038/cddis.2013.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan LP, Seinen E, Duns G, de Jong D, Sibon OC, Poppema S, Kroesen BJ, Kok K, van den Berg A. 2009. A high throughput experimental approach to identify miRNA targets in human cells. Nucleic Acids Res 37:e137. doi: 10.1093/nar/gkp715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nigam R, Laulederkind SJ, Hayman GT, Smith JR, Wang SJ, Lowry TF, Petri V, De Pons J, Tutaj M, Liu W, Jayaraman P, Munzenmaier DH, Worthey EA, Dwinell MR, Shimoyama M, Jacob HJ. 2013. Rat Genome Database: a unique resource for rat, human, and mouse quantitative trait locus data. Physiol Genomics 45:809–816. doi: 10.1152/physiolgenomics.00065.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 55.Dweep H, Sticht C, Pandey P, Gretz N. 2011. miRWalk-database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 56.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2004. Human microRNA targets. PLoS Biol 2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 58.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. 2006. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 59.Kruger J, Rehmsmeier M. 2006. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.GTEx Consortium. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Panwar B, Omenn GS, Guan Y. 2017. miRmine: a database of human miRNA expression profiles. Bioinformatics 33:1554–1560. doi: 10.1093/bioinformatics/btx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leung YY, Kuksa PP, Amlie-Wolf A, Valladares O, Ungar LH, Kannan S, Gregory BD, Wang LS. 2016. DASHR: database of small human noncoding RNAs. Nucleic Acids Res 44(D1):D216–D222. doi: 10.1093/nar/gkv1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koressaar T, Remm M. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 64.Lee C, Huang CH. 2013. LASAGNA-Search: an integrated Web tool for transcription factor binding site search and visualization. Biotechniques 54:141–153. doi: 10.2144/000113999. [DOI] [PubMed] [Google Scholar]
- 65.Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Cheneby J, Kulkarni SR, Tan G, Baranasic D, Arenillas DJ, Sandelin A, Vandepoele K, Lenhard B, Ballester B, Wasserman WW, Parcy F, Mathelier A. 2018. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its Web framework. Nucleic Acids Res 46(D1):D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. 2015. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. 2016. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res 44(D1):D313–D316. doi: 10.1093/nar/gkv1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, Morin PJ, Buetow KH, Strausberg RL, De Souza SJ, Riggins GJ. 2002. An anatomy of normal and malignant gene expression. Proc Natl Acad Sci U S A 99:11287–11292. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. 2018. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 46(D1):D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]