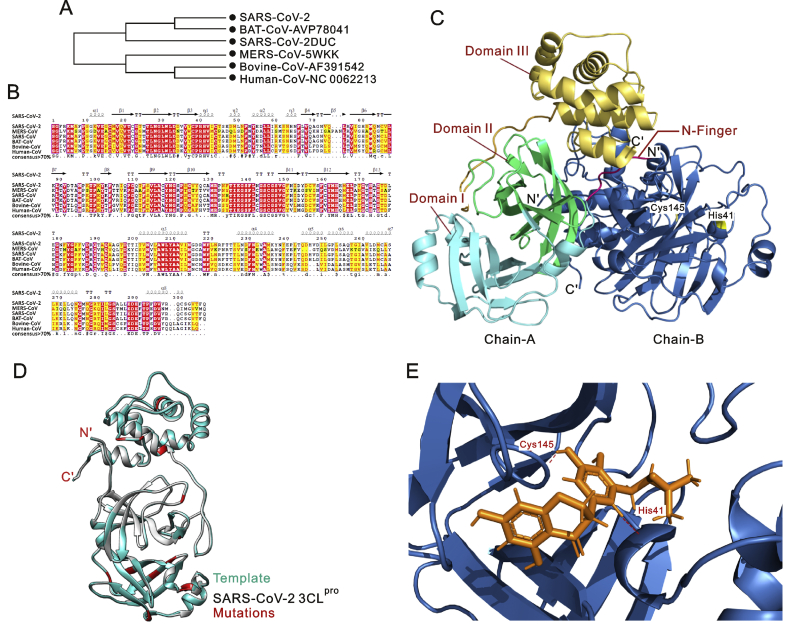
Fig. 1.
(A) Phylogenetic tree inferred from closest homologs of SARS-CoV-2 3CLpro. The maximum likelihood method was used to construct this tree. (B) Multiple sequence alignment of closest homologs of SARS-CoV-2 3CLpro sharing ≥70% sequence identity. (C) Cartoon representation of the SARS-CoV-2 3CLpro homodimer. Chain-A (protomer-A) is in multicolour and Chain-B (protomer-B) is in dark blue. The N-finger that plays an important role in dimerization maintaining the active conformation is shown in hot pink, domain I is coloured cyan, domain II is shown in green, and domain III is coloured yellow. The N- and C-termini are labelled. Residues of the catalytic dyad (Cys-145 and His-41) are highlighted in yellow and labelled. (D) Cartoon representation of the 3CLpro monomer model (chain/protomer-A) of SARS-CoV-2 superimposed with the SARS-CoV 3CLpro structure. The SARS-CoV 3CLpro template is coloured cyan, the SARS-CoV-2 3CLpro structure is coloured grey, and all identified mutations are highlighted in red. (E) Docking of 5,7,3′,4′-tetrahydroxy-2’-(3,3-dimethylallyl) isoflavone inside the receptor-binding site of SARS-CoV-2 3CLpro, showing hydrogen bonds with the catalytic dyad (Cys-145 and His-41). The 3CLpro structure is coloured dark blue, the 5,7,3′,4′-tetrahydroxy-2’-(3,3-dimethylallyl) isoflavone is orange, and hydrogen coloured maroon.