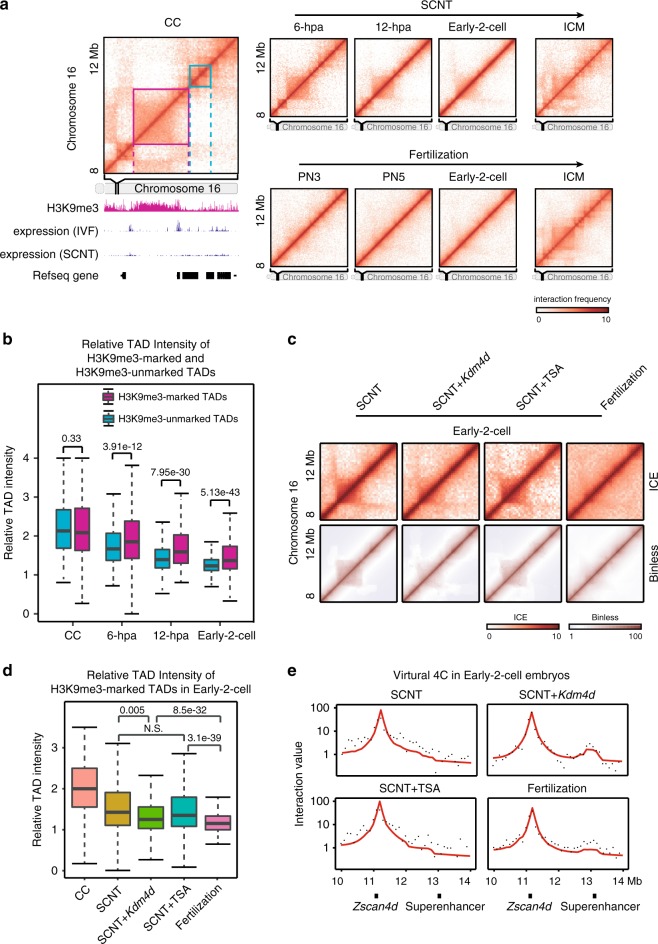
Fig. 6. H3K9me3 is a potential barrier of TADs reprogramming.
a Heatmaps of chromatin interaction frequencies showing a CC TAD unreprogrammed example (magenta) and a CC TAD reprogrammed (cyan) example during SCNT embryo development. The bottom track shows the H3K9me3 signal in CCs. b RTIs of H3K9me3-maked and H3K9me3-unmarked TADs identified in CCs. RTI values in CC, 6-hpa, 12-hpa and early-2-cell embryos are shown (n = 2184). Boxes show 25th, 50th and 75th percentiles and whiskers show 1.5× the inter-quartile range. The two-sided p values were calculated by the Kruskal–Wallis test with Dunn’s multiple comparison test and adjusted by default with the holm method. Source data and exact p-value are provided as a Source Data file. c Heatmaps showing normalized chromatin interaction frequencies (100-kb bin, chromosome 16) in SCNT, SCNT with Kdm4d mRNA injection, SCNT with TSA treatment and fertilization-derived early 2-cell embryos. Both ICE and Binless normalization approaches are used. d RTIs of H3K9me3-maked TAD in early 2-cell embryos. RTI values in CC, SCNT, SCNT with Kdm4d mRNA injection, SCNT with TSA treatment and fertilization-derived are shown (n = 2184). Boxes show 25th, 50th and 75th percentiles and whiskers show 1.5× the inter-quartile range. The two-sided p values were calculated by the Kruskal–Wallis test with Dunn’s multiple comparison test and adjusted by default with the holm method (N.S., not significant). Source data and exact p-value are provided as a Source Data file. e Virtual 4C test showing the interaction frequencies between Zscan4d promoter and its adjacent region in chromosome 7. Black dots represent observed values, red line represents fitted value by Binless.