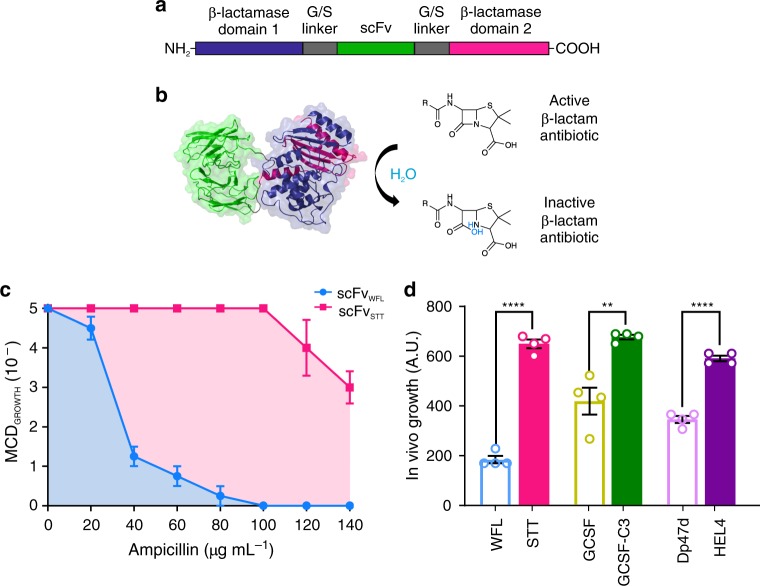
Fig. 1. The tripartite β-lactamase assay.
a The test protein (green) is inserted into a 28-residue glycine/serine-rich linker (grey) separating the two domains of the E. coli enzyme TEM-1 β-lactamase (purple and pink). b Correct folding of the test protein in the E. coli periplasm enables the two halves of β-lactamase to be brought into close proximity to form the functional enzyme active site that hydrolyses β-lactam antibiotics. c Antibiotic survival curve of the maximal cell dilution allowing growth (MCDGROWTH) on solid medium over a range of ampicillin concentrations for bacteria expressing the aggregation-prone scFvWFL within β-lactamase (blue) or the aggregation-resistant sequence scFvSTT (pink). d Calculating the area under the antibiotic survival curves (blue and pink shaded area, c) yields a single value to compare the behaviour of the different sequences. Data are shown for three aggregation-prone model therapeutic proteins (open bars) and their engineered aggregation-resistant counterparts (solid bars). Data represent mean values ± s.e.m. (n = 4 biologically independent experiments). Asterisks denote significance: **p < 0.002, ****p < 0.0001 (two-sided t-test). Source data are provided as a Source Data file.