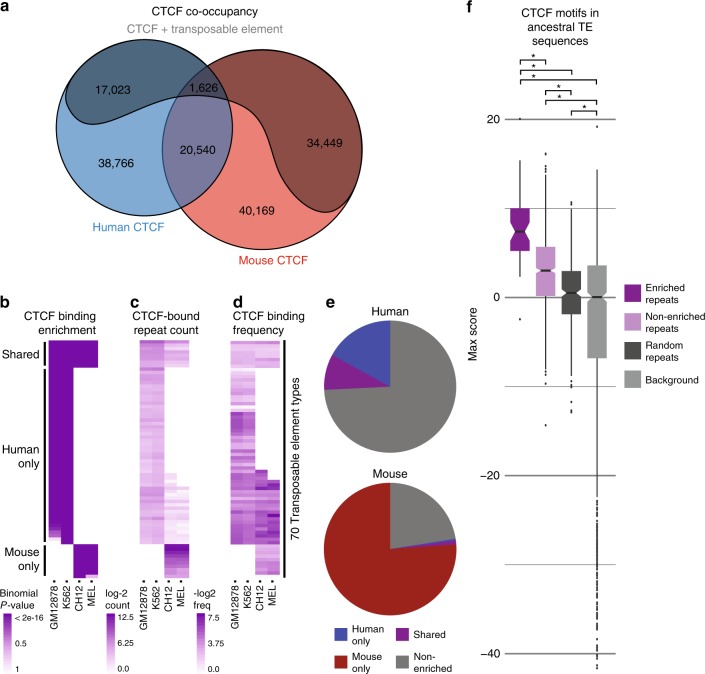
Fig. 2. CTCF-binding variability is associated with transposable element activity.
a Proportion of CTCF sites in the human and mouse genomes with conserved and divergent binding and their respective transposable element (TE)-derived fractions. Human-specific and mouse-specific fractions include both orthologous and non-orthologous CTCF-binding sites. b Binomial tests recovered 70 TE types significantly enriched for CTCF binding. Enrichments were classified as human-only, mouse-only, or shared based on the cell types in which they were observed. c Cell-wise counts of CTCF-bound copies for each enriched TE type. d Cell-wise percentage of TE copies bound by CTCF for each enriched TE type. e Human and mouse fractions of TE-derived CTCF-binding sites originating from human-enriched, mouse-enriched, shared, and non-enriched TE types. f Log-odds score distributions for the strongest CTCF motif match within consensus of CTCF-enriched and non-enriched TEs, compared with TEs selected randomly from RepBase and length-matched background sequences. Scores above 1 represent sequences with greater than random resemblance to the CTCF motif. Enriched repeats, n = 53; non-enriched repeats, n = 905; random repeats, n = 343; background, n = 958. Boxplots are centered around the median, with upper and lower hinges indicating the first and third quartiles. Upper and lower whiskers extend from the hinge to the largest and smallest values within 1.5× the inter-quartile range from the hinge. Individual data points beyond the ends of the whiskers represent outliers. *One-sided Wilcoxon rank-sum test p-value < = 0.03.