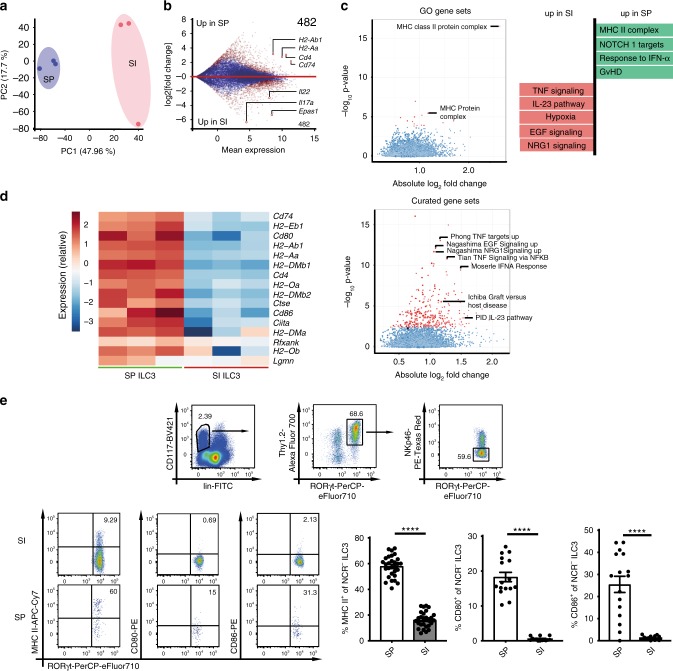
Fig. 1. SP and SI NCR− ILC3s exhibit a different transcriptional signature.
a PCA of RNA sequencing data of SI and SP NCR− ILC3s isolated from Rorγtfm+ mice. Cells were sort-purified as depicted in Supplementary Fig. 1a. b Mean expression and log 2(fold change) of all detected genes. Genes with a significant difference are highlighted in red (FDR < 0.05). Numbers indicate the total amount of genes significantly higher expressed (log2(fold change)>1.5) in SP ILC3s or SI ILC3s. c Gene set enrichment analysis of gene ontology (GO) and curated gene sets. Gene sets with a significant difference are highlighted in red (FDR < 0.05). d Heatmap of genes associated with MHC II Ag presentation. e CD117+lin−Thy1.2+RORγt+NKp46− ILC3s were analyzed for surface expression of MHC II (n = 29(SP) and n = 27(SI) mice), CD80 (n = 16(SP) and n = 14(SI) mice), and CD86 (n = 16(SP) and n = 14(SI) mice). Six to eight independent experiments. Each symbol represents a sample and the bar graph represents the mean ± s.e.m. ****P ≤ 0.0001, calculated with unpaired two-tailed Student’s t test. Source data are provided as a Source Data File.