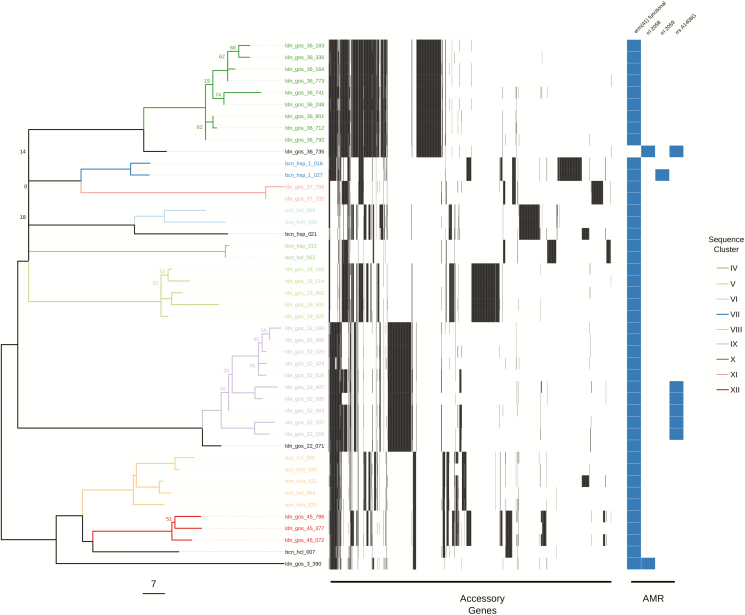
Figure 4.
Maximum likelihood SNV tree for all ST-1 isolates. SNVs were identified from mapping reads to Mycobacterium abscessus subsp. abscessus ATCC19977. Samples are highlighted based on inclusion in sequence clusters. The tree is annotated with the presence (black) and absence (white) of accessory genes, as well as the presence of AMR-associated genes and mutations. This included the presence of a functional erm(41) gene conferring inducible resistance to macrolides; the presence of 2 rrl mutations conferring high-level macrolide resistance; and the presence of a mutation in rrs conferring high-level amikacin resistance. The scale bar represents the number of SNVs, and the node bootstrap scores below are shown if below 75. Abbreviations: AMR, antimicrobial resistance; SNV, single nucleotide variant.