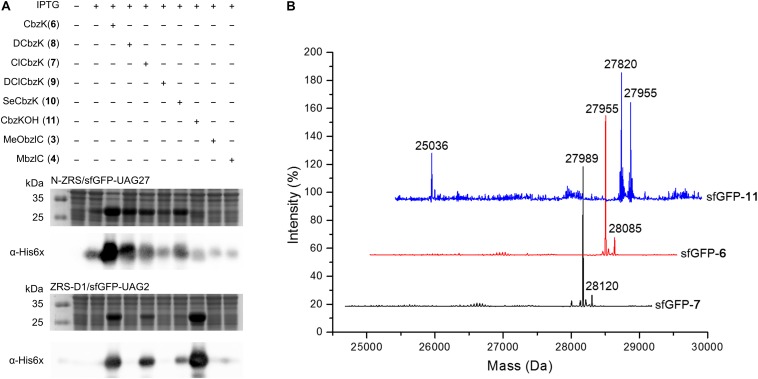
FIGURE 6.
sfGFP production by ZRS variants and mass characterization. (A) Amber suppression of the sfGFP-UAG27 gene (N-ZRS⋅tRNAPyl pair) and the sfGFP-UAG2 gene (ZRS-D1⋅tRNAPyl pair) product with ncAA 3–4 and 6–11. The sfGFP proteins were produced in E. coli BL21 (DE3) coding N-ZRS⋅tRNAPyl or ZRS-D1⋅tRNAPyl pair with the supplement of 1 mM IPTG and ncAAs in GMML medium at 37°C for 12 h. The whole-cell lysate was analyzed by SDS-PAGE and western blotting by anti-His tag antibody indicated as α-His6X. (B) ESI-MS determination of sfGFP-UAG27 proteins with ncAA 6, 7, and 11. Full-length sfGFP-6 and sfGFP-7 proteins were produced by N-ZRS⋅tRNAPyl pair in E. coli BL21 (DE3) with the supplement of 1 mM IPTG and ncAA 6 or 7 in LB medium at 37°C for 12 h. Full-length sfGFP-11 proteins were produced with the same condition but with 1 mM ncAA 11 and in GMML minimal medium. The calculated molecular masses of sfGFP-7 are 28,121 and 27,989 Da (–Met); the observed molecular masses are 28,120 and 27,989 Da (–Met). The calculated molecular masses of sfGFP-6 are 28,085 and 27,954 Da (–Met); the observed molecular masses are 28,085 and 27,955 Da (–Met). The calculated molecular masses of sfGFP-11 are 28,086, 27,955 (–Met), and 25,320 Da (truncated sfGFP at 27 position); the observed molecular masses are 27,955 (–Met) and 27,820 Da (without Cbz group at 27 position and N-terminal Met residues) and 25,036 Da. The detailed electrospray and deconvoluted mass spectra are shown in Supplementary Figures S4–S6. ESI-MS determination of sfGFP-UAG2 protein. sfGFP-11* with ncAA 11 (ZRS-D1⋅tRNAPyl pair) is shown in Supplementary Figure S7.