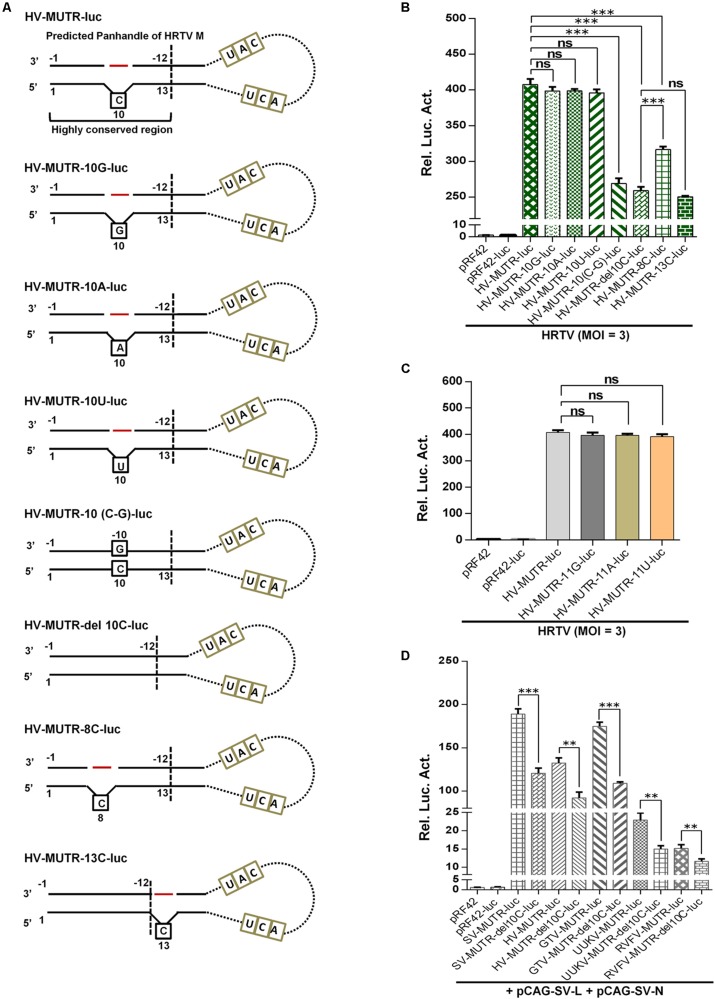
FIGURE 7.
The conserved protruding nucleotide of the panhandle structure is important for M genome UTR activity. (A) Schematic of HRTV M minigenomes with the protruding nucleotide mutated. HV-MUTR-10G, HV-MUTR-10A, and HV-MUTR-10U refer to the mutants of which the protruding 10C in 5′ UTR was mutated to G, A, and U, respectively. HV-MUTR-10(C-G), the mutant with base-pairing C-G at positions 10 of 5′ UTR and –10 of 3′ UTR by inserting a G into 3′ terminal correspondingly. HV-MUTR-del10C, the mutant with the protruding 10C deleted. HV-MUTR-8C and HV-MUTR-13C, the mutants in which a C was respectively introduced at position 8 or 13 of 5′ UTR based on the mutant “HV-MUTR-del10C” by PCR. (B) BHK-21 cells transfected with the indicated control or minigenome luc plasmids and pRL-TK were superinfected with HRTV (MOI = 3) at 12 h post-transfection, followed by luc activity measurements at 48 hpi. (C) Mutants with the protruding 11C changed to G, A, or U were also similarly constructed. Superinfection-based minigenome reporter assays were conducted as in (B). (D) Minigenome-luc plasmids for M segments of GTV (GV-MUTR-luc), UUKV (UV-MUTR-luc), and RVFV (RV-MUTR-luc) were similarly generated by cloning the viral M minigenome-luc chimeras into pRF42 and based on these wild type minigenomes, the corresponding mutants with the protruding 10C deleted were further constructed. BHK-21 cells were co-transfected with the indicated minigenome-luc plasmids or the controls, together with the SFTSV N and L expression plasmids and pRL-TK. At 48 h post-transfection, luc activities were measured. Data shown are mean ± SEM, n = 3. ∗∗P < 0.01; ∗∗∗P < 0.001; ns, non-significant.