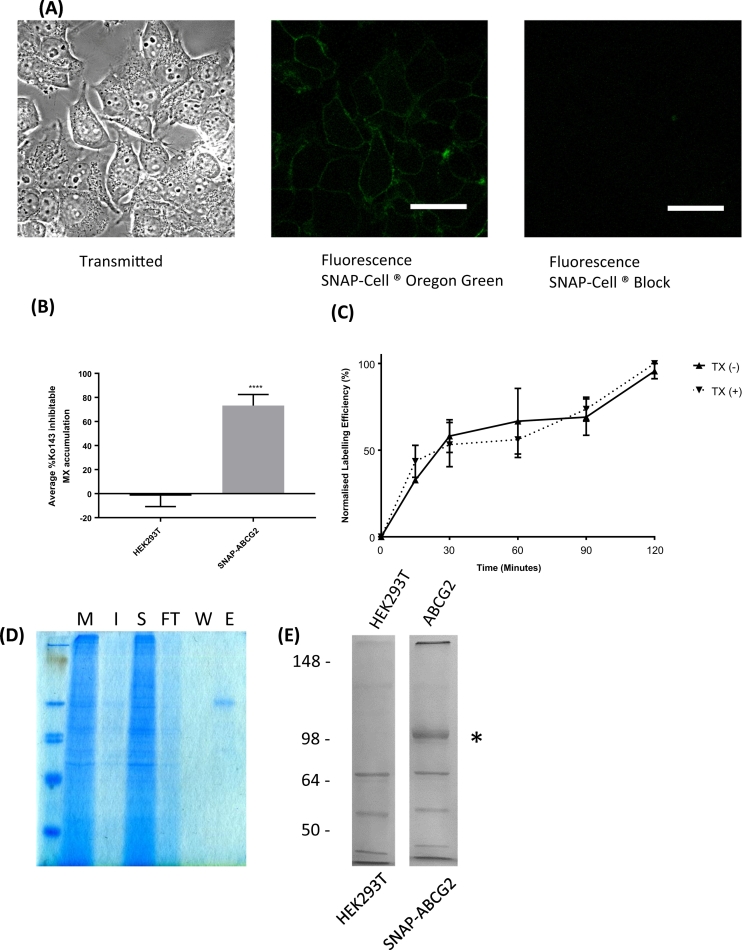
Fig. 4.
SNAP-ABCG2 surface expression, activity and purification. SNAP-ABCG2 expressing cells were labelled for 30 min at 37 °C 5% CO2 with 1 μM SNAP-Cell Oregon Green alone (middle) or after pre-incubation with 2 μM SNAP-Cell ® Block (right panel). After washing cells were imaged using an LSM710 confocal microscope (Carl Zeiss) with fluorescence images gathered using 488 nm/493-598 nm excitation/emission wavelengths. Scale bar = 20 μm. (B) Corrected mitoxantrone (MX) fluorescence intensity values were compared in the presence and absence of Ko143 and the function of ABCG2 was determined as % Ko143 inhibitable MX accumulation. Data are plotted as mean ± SD (n = 4) with statistical significance (****, p < 0.001) compared to parental HEK293T assessed by unpaired t-test. (C) Labelling of SNAP-ABCG2 in membrane fractions prior to purification. Membranes were incubated with SNAP-Surface AlexaFluor® 647 in the presence (+) of absence (−) of 0.5% v/v Triton X-100 for the indicated times and fluorescence measured with a fluorimeter. (D) Purification of SNAP-ABCG2 by metal affinity chromatography, following SMALP solubilisation. Fractions indicate whole cell membranes (M), SMALP-insoluble (I), SMALP-soluble (S), flow through (FT), wash (W) and elution (E). (E) Purification fractions containing purified SNAP-ABCG2 were concentrated by centrifugation. A preparation of material from cells not expressing ABCG2 was treated similarly. 10 μL of each of these samples was run on an 8% (w/v) polyacrylamide gel and stained InstantBlue. ABCG2 (identified *) and some contaminants were revealed.