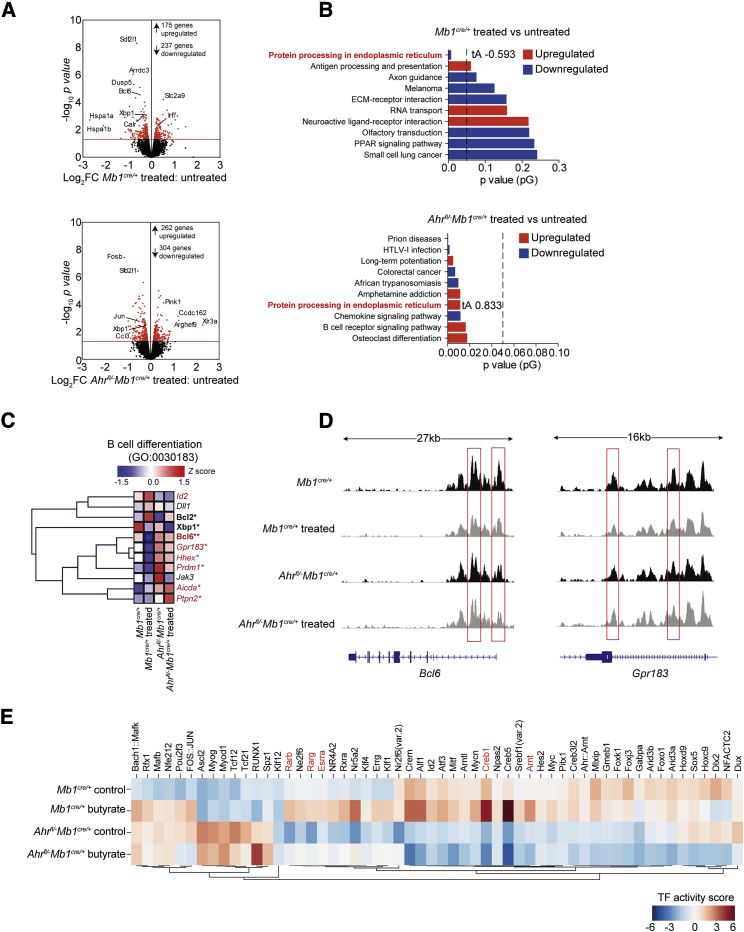
Figure 4.
Butyrate Supplementation Modulates the Transcriptional and Epigenetic Landscape of CD19+CD21hiCD24hiB Cells in an AhR-Dependent Manner
(A) Volcano plots shows log2 fold change (FC) in gene expression between CD19+CD21hiCD24hiB cells isolated from butyrate-supplemented Mb1cre/+ mice compared to control Mb1cre/+ mice (top plot) and between butyrate supplemented Ahrfl/−Mb1cre/+ compared to control Ahrfl/−Mb1cre/+ mice (bottom plot). Red dots represent significant DEGs, with the red line denoting a cut off p value of <0.05.
(B) Signaling pathway impact analysis (SPIA) ranked on significance (pG) comparing the over-represented (red) and under-represented (blue) pathways in butyrate-supplemented compared to control CD19+CD21hiCD24hiB cells from Mb1cre/+ mice (top graph) and Ahrfl/−Mb1cre/+ mice (bottom graph). The total perturbation accumulation (tA) score is listed for the “protein processing in endoplasmic reticulum” pathway.
(C) Heatmap shows the expression of B cell differentiation genes in CD19+CD21hiCD24hiB cells isolated from control Mb1cre/+ mice, butyrate-supplemented Mb1cre/+ mice, control Ahrfl/−Mb1cre/+ mice, and butyrate-supplemented Ahrfl/−Mb1cre/+ mice. Mean z scores were calculated from log CPM values. Samples highlighted in red are significantly differentially expressed between CD19+CD21hiCD24hiB cells isolated from butyrate-supplemented Mb1cre/+ mice compared to butyrate-supplemented Ahrfl/−Mb1cre/+ mice. Samples highlighted in bold are significantly differentially expressed between CD19+CD21hiCD24hiB cells isolated from butyrate-supplemented Mb1cre/+ mice compared to control Mb1cre/+ mice.
(D) Representative ATAC-seq tracks for the Bcl6 and Gpr183 loci in CD19+CD21hiCD24hiB cells from butyrate-supplemented or control Mb1cre/+ and Ahrfl/−Mb1cre/+ mice (n = 3). Track heights between samples are normalized through group autoscaling. For RNA-seq data, n = 3 per condition and genotype.
(E) Heatmap shows inferred transcription factor activity scores based on accessibility at transcription factor binding motifs in CD19+CD21hiCD24hiB cells isolated from control Mb1cre/+ mice, butyrate-supplemented Mb1cre/+ mice, control Ahrfl/−Mb1cre/+ mice, and butyrate-supplemented Ahrfl/−Mb1cre/+ mice as measured by ATAC-seq. AhR co-factors are highlighted in red. For ATAC-seq data, n = 3 for Mb1cre/+ mice and n = 2 for Ahrfl/−Mb1cre/+ mice. For RNA-seq data, n = 3 per group.
Cells were isolated at day 7 post-disease onset. See also Figure S7.