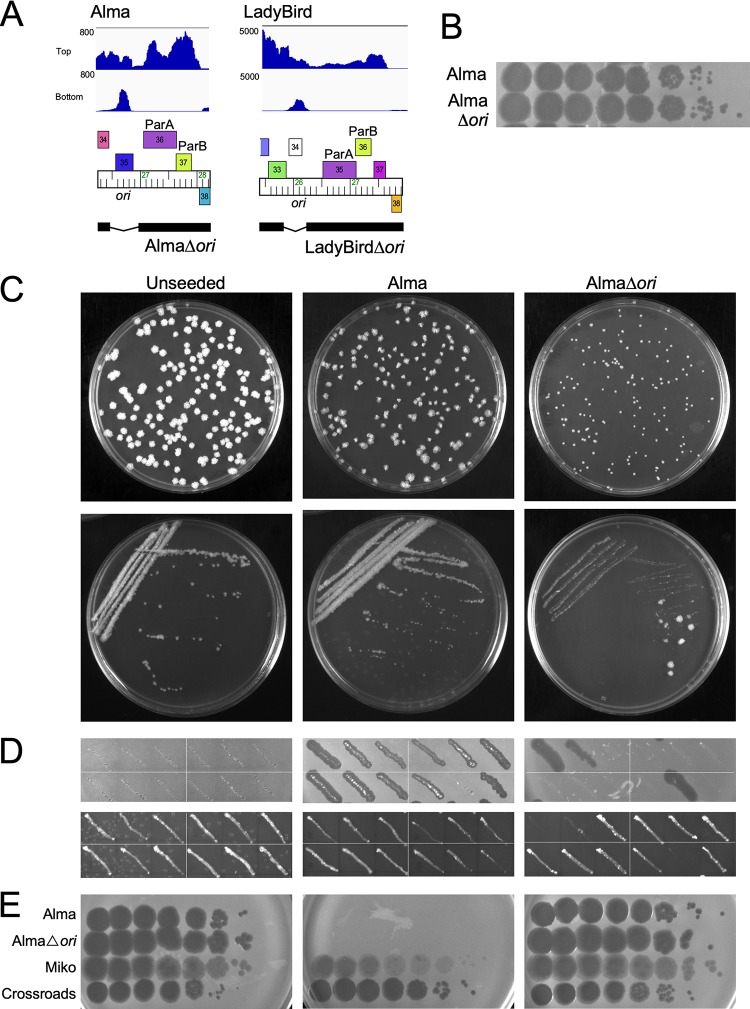
FIG 4.
ori is necessary to form stable lysogens of Alma. (A) Genomes of Δori mutant phages. Phages were engineered to delete the noncoding RNA region (labeled ori) expressed by the prophage (Alma, LadyBird). The black bar below the genome map segment shows the retained regions. The coordinates of the deleted regions are 26463 to 26959 and 25900 to 26299 for Alma and LadyBird, respectively. Strand-specific RNA-Seq reads are aligned above the Alma and LadyBird maps. (B) Titer and plaque morphology of AlmaΔori. Lysates of Alma and AlmaΔori were 10-fold serially diluted and plated onto a lawn of M. smegmatis mc2155. (C) Characterization of AlmaΔori lysogens (akin to Fig. 3C). Lysogens were recovered by plating serial dilutions of exponentially growing M. smegmatis mc2155 onto unseeded solid media or solid media seeded with Alma or AlmaΔori (Top). Ten (Alma, AlmaΔori) or six (unseeded) individual colonies were streaked onto solid media to remove phage particles carried over from the selection plate (bottom) (representative shown). (D) Spontaneous phage release from colonies grown in the presence of AlmaΔori. Three colonies from each streak plate were patched onto solid media and M. smegmatis lawns to test for spontaneous phage release. None of the colonies recovered on unseeded media released phage, but at least two of the three colonies from purified streaks from colonies recovered on Alma-seeded plates are lysogenic and release phage particles. Some patches originating from AlmaΔori-seeded plates released phage, while others did not. Patches that did not release phage grew well on solid media, while patches that did release phage grew poorly on solid media. (E) Susceptibility of colonies to phage superinfection. Liquid cultures were grown from patches of 6 of the 10 colonies from the seeded plates and tested for their susceptibilities to Alma, AlmaΔori, Miko and a control phage Crossroads (L2). A representative example of each is shown.