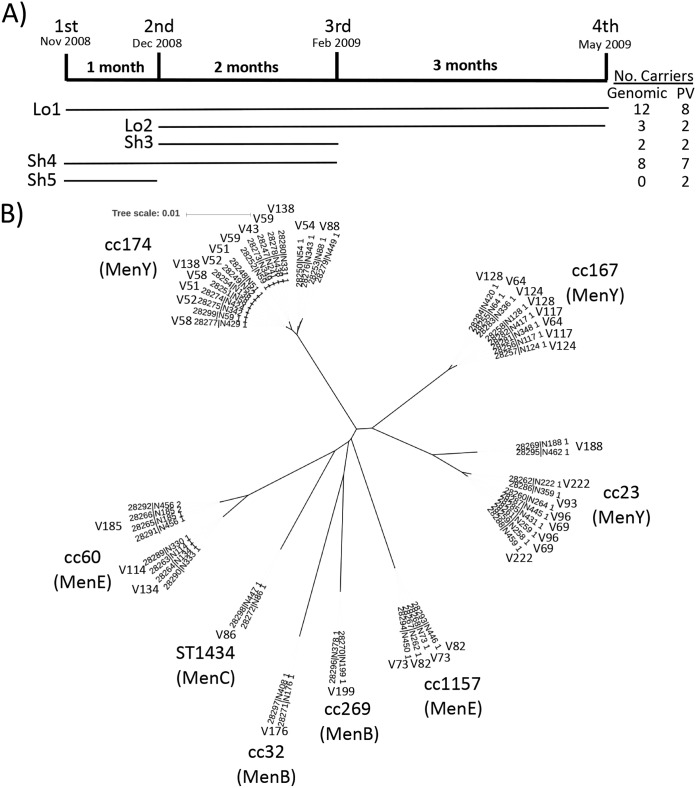
FIG 1.
Core genome phylogenetic tree for 25 pairs of short- and long-term persistent carriage isolates. Persistent carriage isolates were chosen from volunteers, all first-year students that were part of a longitudinal carriage study at the University of Nottingham. For 25 volunteers, one isolate was chosen from the first and last time point where persistent carriage was observed and subject to WGS by Illumina HiSeq. Analysis of the PV states of 14 SSR-containing genes was also performed on multiple isolates for 21 volunteers. (A) Sampling times, the distribution of carriers with respect to long-term (Lo; 5 to 6 months) and short-term (Sh; 1 to 3 months) carriage, and the number of carriers whose isolates were subject to WGS (genomic) and/or PV analysis. (B) Phylogenetic tree derived using a subset of the Neisseria core genes (n = 215) and iTOL from whole-genome sequences generated on an Illumina HiSeq platform. The specific volunteers (see Table 1 for isolate names for each volunteer) in each group are: Lo1, V51, V58, V59, V73, V86, V88, V117, V128, V176, V185, V188, and V222; Lo2, V69, V82, and V96; Sh3, V43 and V93; Sh4, V52, V54, V64, V114, V124, V134, V138, and V199; Sh5, V113, and V115. Individual branches of the tree are labeled with the clonal complex (cc) and genogroup (capsule type). The volunteer for each isolate is also indicated (there is a single label where two or more isolates are on adjacent branches).