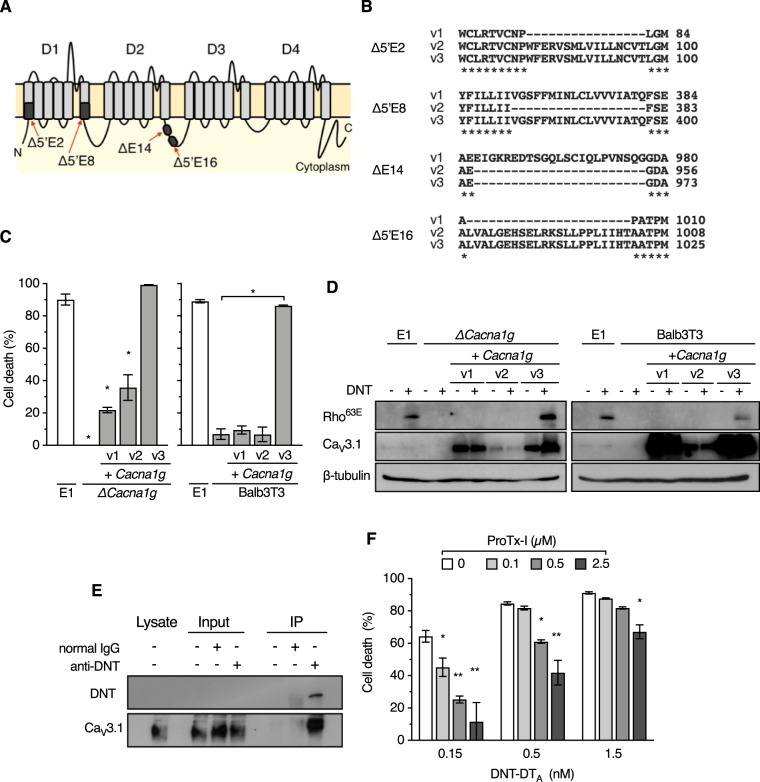
FIG 3.
Identification of CaV3.1 as a receptor for DNT. (A) Schematic illustration of CaV3.1 consisting of four distinct domains (D1 to D4). Missing regions by alternative splicing in the variants are indicated by black boxes. Each region is designated according to the designations in a previous study (24). (B) Amino acid sequence alignments of the spliced regions of v1, v2, and v3. Dashes indicate missing regions of the splice variants. Asterisks indicate conserved amino acid residues. (C) Ectopic expression of Cacna1g v3 restores the sensitivity of MC3T3-E1/ΔCacna1g and Balb3T3 cells to DNT-DTA. MC3T3-E1, MC3T3-E1/ΔCacna1g (left), and Balb3T3 (right) cells with or without Cacna1g complementation were treated with DNT-DTA. Each bar represents the mean ± SEM (n = 3). *, P < 0.0001 (compared with E1 [left] and Balb3T3 [right]). (D) Ectopic expression of Cacna1g v3 restores the sensitivity of MC3T3-E1/ΔCacna1g (left) and Balb3T3 (right) cells to DNT. The cells were treated with DNT and subjected to SDS-PAGE, followed by immunoblotting. (E) Immunoprecipitation (IP) assay to detect interactions between CaV3.1 and DNT. After treatment with DNT, MC3T3-E1/ΔCacna1g/+Cacna1g v3 cells were subjected to an immunoprecipitation assay, followed by immunoblotting with anti-DNT antibody and anti-CaV3.1 antibody. (F) Competitive inhibition of the cytotoxicity of DNT-DTA with ProTx-I. MC3T3-E1 cells were treated with DNT-DTA at the indicated concentrations in the presence of ProTx-I and subjected to a cytotoxicity assay. Each bar represents the mean ± SEM (n = 3). *, P < 0.05; **, P < 0.0001 (compared with 0 μM ProTx-I in each DNT-DTA dose group).