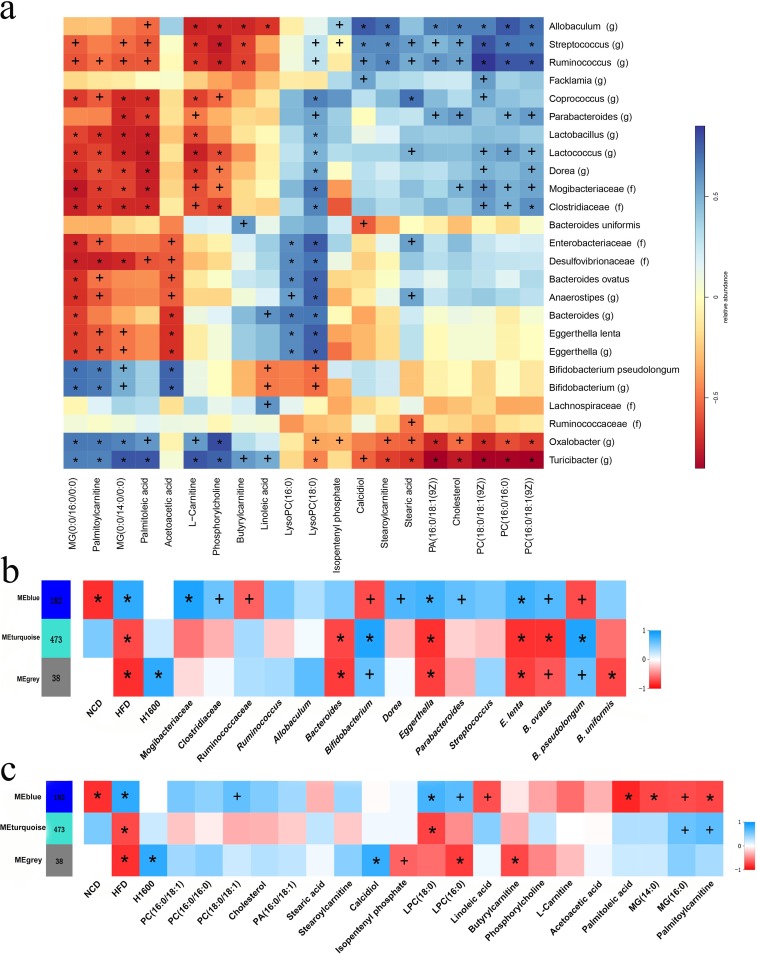
FIG 8.
Correlation analyses of serum metabonomics, gut microbiomes, and hepatic transcriptomes. (a) Spearman correlation matrix of the gut microbiota at the genus and species levels and serum metabolites participating in lipid metabolism (the significantly changed bacterial taxa and serum metabolites that were identified by a nonparametric factorial Kruskal-Wallis test and unpaired Wilcoxon comparison test are represented). A parenthetical “g” or “f” indicates genus or family, respectively. (b) A Spearman correlation matrix of a WGCNA identified clusters in hepatic DEGs between the GML-treated group and the HFD controls and gut microbiota at the genus and species levels (only the significantly changed bacterial taxa that were identified by a nonparametric factorial Kruskal-Wallis test and unpaired Wilcoxon comparison test are represented). ME, methylene. (c) A Spearman correlation matrix of WGCNA identified clusters in hepatic DEGs between the GML-treated group and HFD controls and serum metabolites participating in lipid metabolism (only the significantly changed metabolites that were identified by a nonparametric factorial Kruskal-Wallis test and unpaired Wilcoxon comparison test are represented). Six (a) and four (b, c) mice were tested. Significant correlations are marked by + (P < 0.05) and * (P < 0.01).