Figure 1.
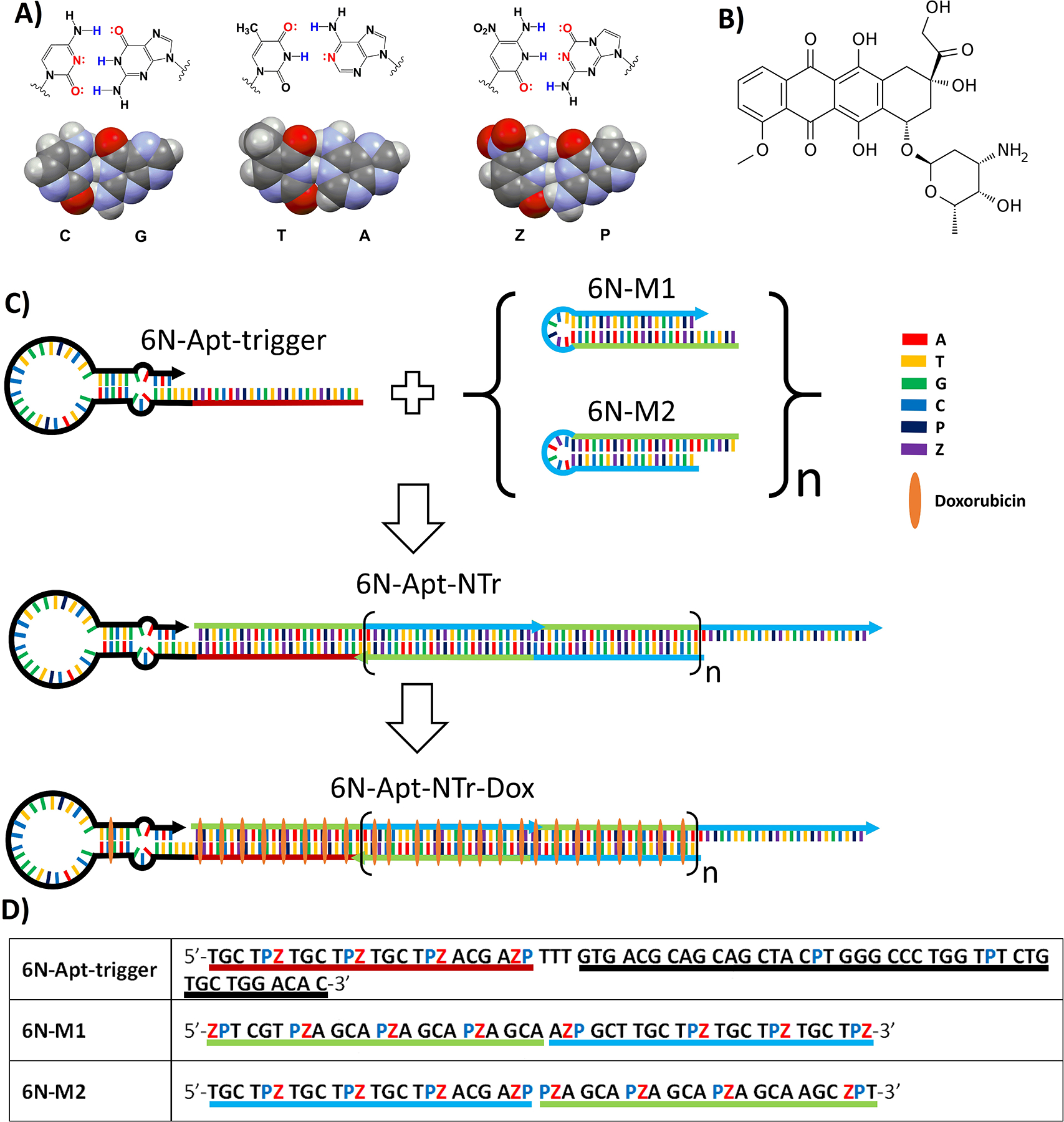
Key components of AEGIS aptamer-nanotrain assembly. (A) Structures of three nucleobase pairs (G:C, A:T, and P:Z) that support nanoassembly in GACTZP 6-nucleotide DNA (PDB ID: 4RHD) (B) Doxorubicin. (C) Self-assembling nanostructure triggered by an aptamer (6N-Apt-trigger) that hybridizes to the toehold of 6N-M1, invades its hairpin to generate a single strand that can invade the hairpin of 6N-M2. This yields a 6-nucleotide DNA nanotrain (6N-Apt-NTrs) formed from short 6-nucleotide DNA fragments (6N-M1 and 6N-M2) tethered to a 6-nucleotide aptamer specific for liver cancer cells. The nanotrain may be loaded with the doxorubicin drug. (D) Sequences of 6-nucleotide DNA fragments. Underlined region highlighted with red color shows the sequence of the trigger; Black color underlined region shows the sequence of the aptamer; Green and blue color underlined regions in 6N-M1 and 6N-M2 show the complementary regions when forming nanotrains.
