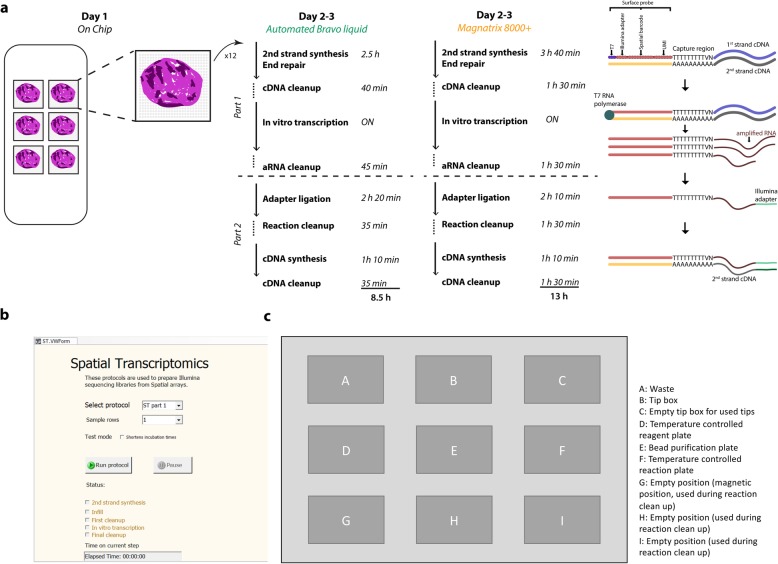
Fig. 1.
Workflow for automated ST library preparation. a Each ST barcoded array contains six subarrays, each with 2000 100-μm spots. Every spot contains oligo-dT probes bearing a spot-specific barcode. The protocol is divided into four parts. The first is performed on the chip, where fresh frozen tissue sections are mounted on the barcoded subarrays. The tissue sections are permeabilized, allowing their mRNA to be captured by the oligo-dT probes on the surface, which function as primers for overnight cDNA synthesis. On the following day, the tissue sections are removed from the subarray surface. The cDNA-mRNA hybrids are then released and collected per sample and transferred to the Bravo system for the second and third parts of the protocol. Finally, the libraries undergo PCR indexing in parallel before sequencing. b Graphical interface to the automated program. c Layout of the Bravo working deck prior to start. Positions A to C are used for tips and waste, while the reaction plate, containing the input material together with master mixes for the enzymatic reactions is placed on position D, which is kept at 4 °C. On position E, a 2 mL deep well plate is holding the reagents for the bead clean up steps. An empty 96-well plate is placed on the temperature controlled position F, which is where the enzymatic reactions are carried out. Positions G to H are used during reaction clean up