Figure 1. Post-transcriptional mechanisms influence proteome content.
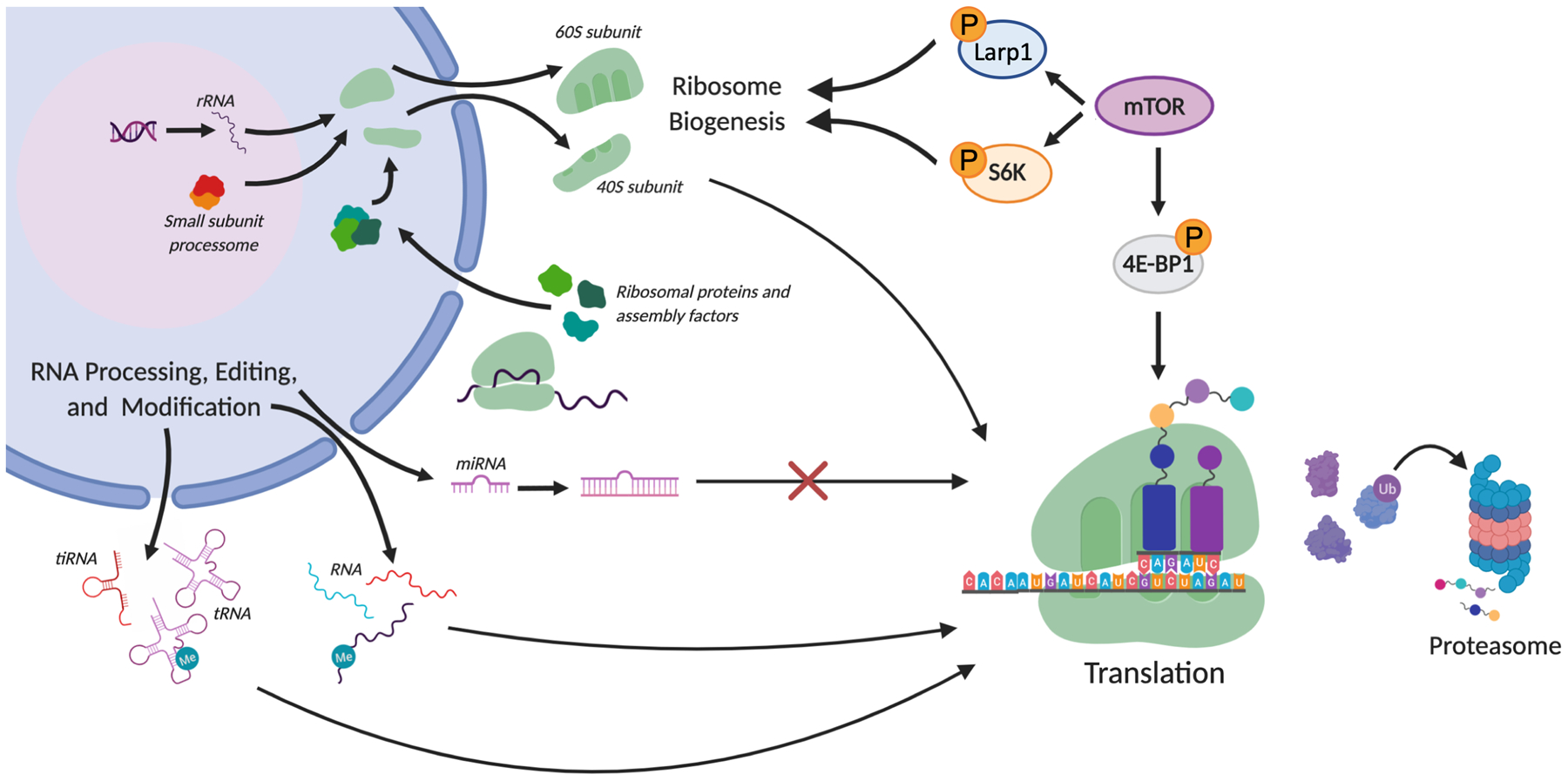
Proteome content can be regulated by RNA processing, ribosome biogenesis, signaling pathways and protein degradation. RNA splicing can produce mRNAs that code for distinct protein isoforms, introduce premature stop codons, or cause UTR variation that alters translational efficiency. mRNA methylation can alter transcript stability, localization and translational efficiency. Methylation and pseudouridylation of rRNA and tRNA can impact ribosome biogenesis, polysome assembly, translation fidelity and tRNA stability. RNA editing can alter miRNA biogenesis and alter mRNA coding or UTR sequence to alter translational efficiency. The mTOR signaling pathway can promote protein synthesis by enhancing the translation of ribosomal protein mRNAs via phosphorylation of Larp1, ribosome biogenesis via phosphorylation and activation of S6K, and translation initiation via phosphorylation and inhibition of 4E-BPs. The ubiquitin-proteasome system is a major cellular degradation system that contributes to maintaining proteostasis in stem cells by regulating both the content and quality of the proteome through the normal turnover of proteins and the degradation of misfolded proteins.
