Figure 1 |. The ICeChIP-seq workflow.
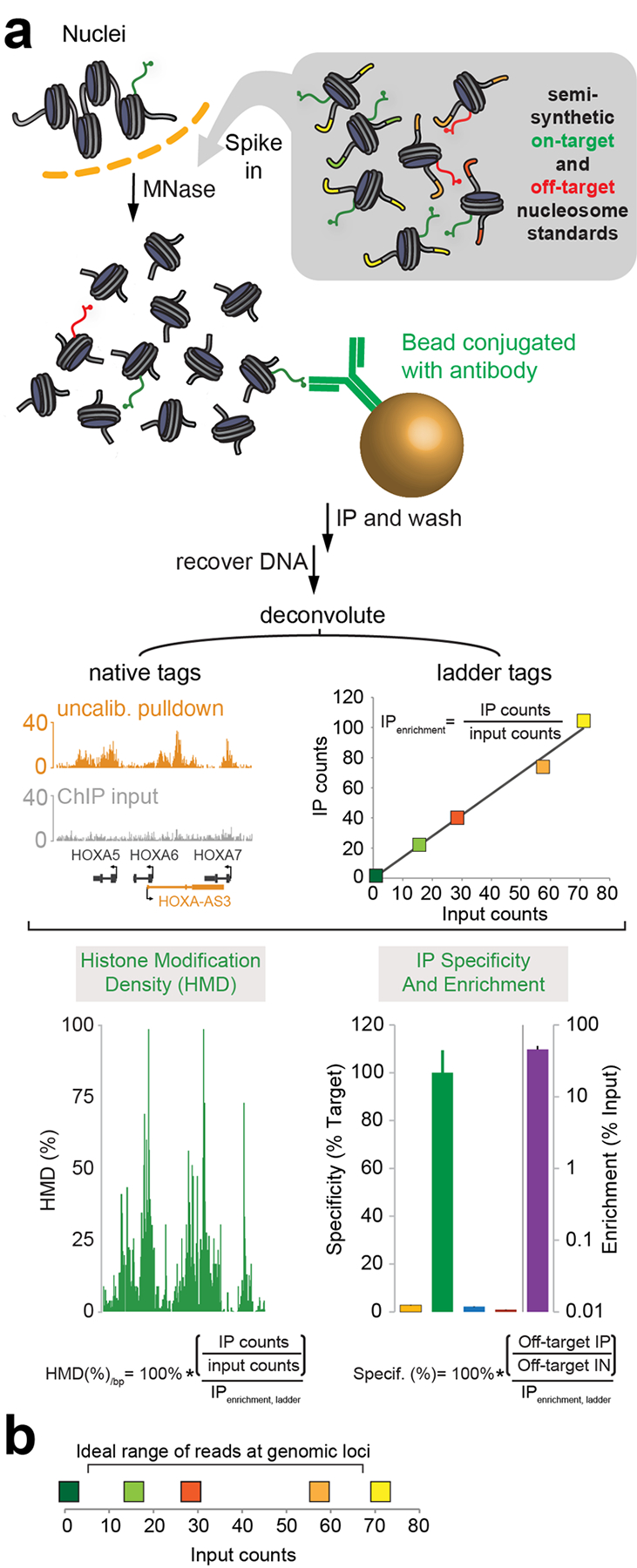
(a) Purified nuclei are spiked with barcoded nucleosome standards representing on- and off-target modifications before chromatin is fragmented by MNase digestion. Fragmented chromatin is then incubated with antibody conjugated to magnetic beads to purify targeted nucleosomes. DNA is then recovered, sequenced, and mapped to the genome or to nucleosome standard barcodes. Histone modification density can be computed as the ratio of locus enrichment to on-target enrichment, and antibody specificity can be computed as the ratio of off-target to on-target enrichment. Adapted from Grzybowski et al.43 and Shah et al.45 Error bars represent standard deviation, as described in the original publications. (b) Example scatterplot showing reads from genomic loci falling between the reads from the highest- and lowest-abundance ladder members for optimal quantification. Ideally, the range of the reads from the barcodes on the target modification standard should encompass the range of input counts at the genomic loci. Different colors each refer to different ladder members comprising the nucleosome ladder.
