Figure 2 |. ICeChIP-seq Bioinformatics Analysis Pipeline.
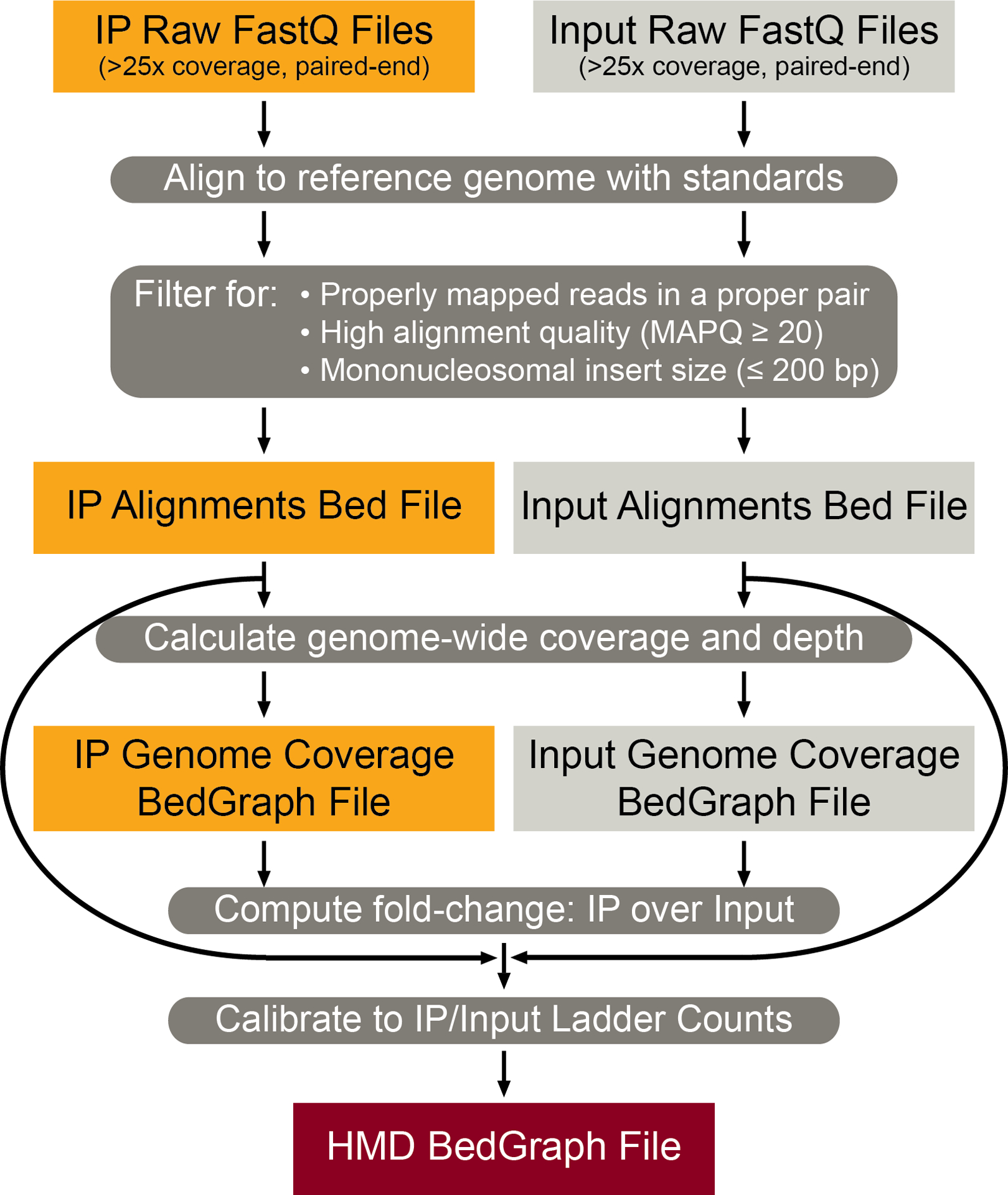
Workflow for analysis of ICeChIP-seq data. IP and Input FastQ paired-end reads are mapped and filtered for quality and mononucleosomal size into their respective alignment Bed files. From these Bed files, genome-wide coverage BedGraph files are generated and merged to compute the fold change of the IP over Input at genomic loci as a BedGraph file. This ratiometric BedGraph file is then calibrated to the nucleosome standard enrichment to generate the final HMD BedGraph file.
