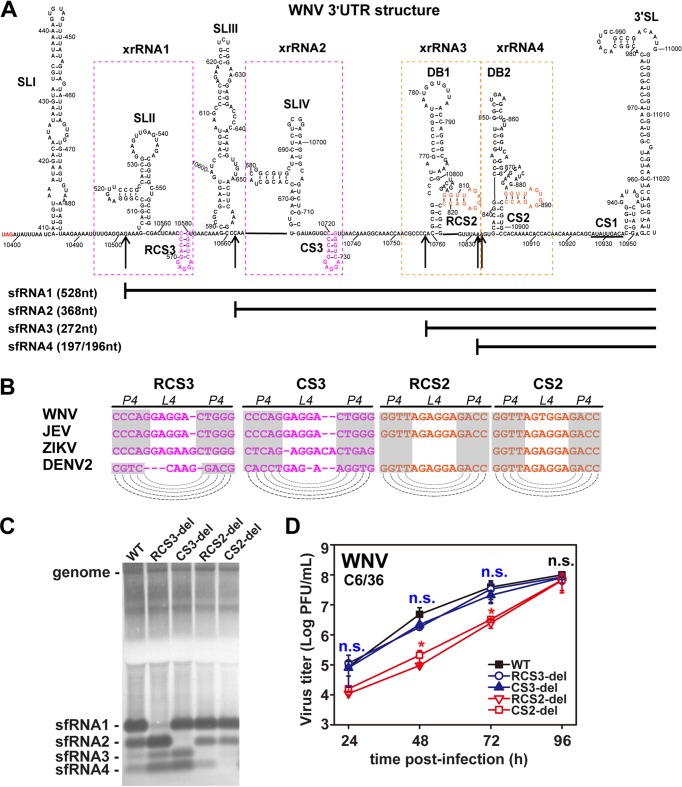
FIG 5.
Conserved DRs of WNV control production of various sfRNA species. (A) Secondary structure of WNV 3ʹ-UTR. The various secondary structures formed within the 3ʹ-UTR are indicated. The putative SLs of RCS3/CS3 and RCS2/CS2 are marked in pink and orange, respectively. The 5′ ends of sfRNA1, sfRNA2, sfRNA3, and sfRNA4 are indicated by arrows. The sizes of different sfRNA species are also indicated. xrRNA1, xrRNA2, xrRNA3, and xrRNA4 are depicted by dotted boxes. (B) Conserved repeated sequences from various flaviviruses. Dashed lines indicate putative base pairing with the SL structures of conserved repeated sequences as indicated in the diagram. (C) Northern blot of total RNA extracted from C6/36 cells infected with WT or mutant WNVs with RCS3, CS3, RCS2, or CS2 deletions. sfRNA signals are marked as sfRNA1, sfRNA2, sfRNA3, and sfRNA4 in descending size order. (D) Growth kinetics of recombinant WNVs with individual RCS3, CS3, RCS2, and CS2 deletions in C6/36 cells. RCS3-del, CS3-del, and WT virus titers showed no significant differences at different time points. n.s., no statistically significant differences. RCS2-del and CS2-del virus titers showed statistically significant differences from the WT virus titers at 24, 48, and 72 h postinfection (hpi) (*, P < 0.05), and there were no statistically significant differences at 96 hpi.