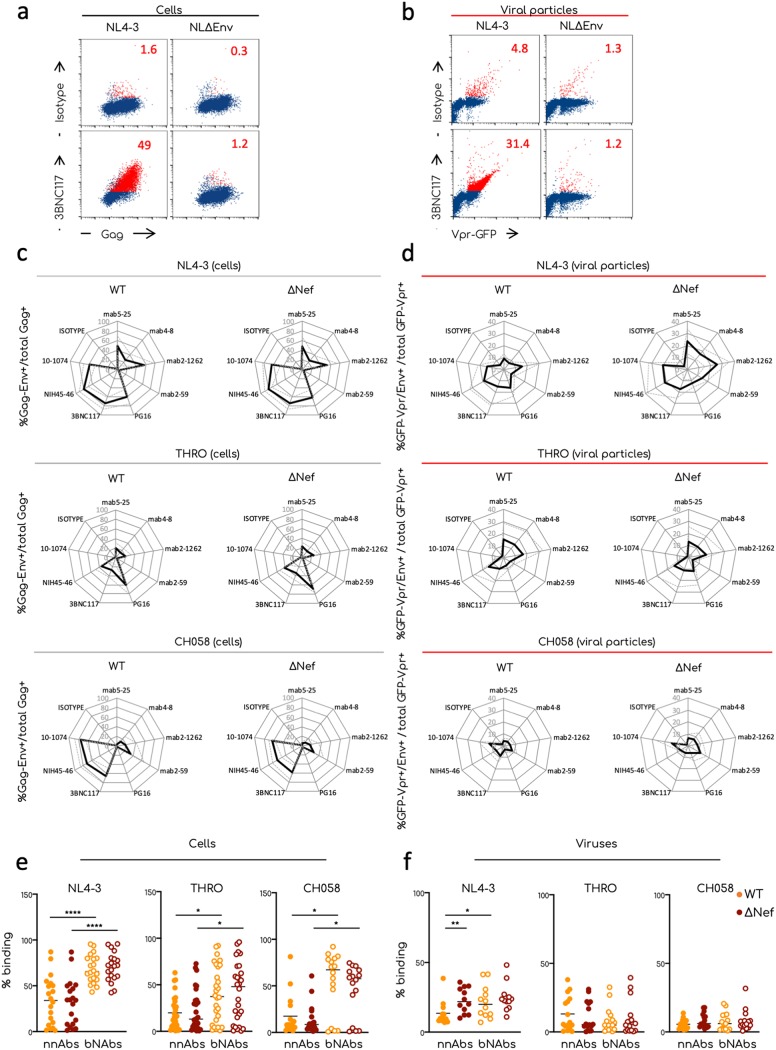
FIG 4.
Comparative analysis of Env conformation at the surface of producer cells and viral particles. Transfected 293T cells and aggregated viral particles were stained for the expression of Env using the indicated antibodies. (a) Representative dot plot analyses of 293T cells transfected with the NL4-3-WT or -ΔEnv proviruses and stained for Gag and Env expression. 293T cells were plotted as Gag+/Env+ events. Numbers indicate the percentages of Gag/Env double-positive events (in red in the plots). (b) Representative dot plot analysis of viral aggregates stained to visualize Env at the surface of the virions. Numbers indicate the percentages of GFP-Vpr/Env double-positive events (in red in the plots). (c and d) Radar plot analysis showing the means (plain lines) and standard deviations (SD; dotted lines) of the Env profiles of the indicated transfected cells (c) and viral aggregates (d). The percentages were calculated by flow cytometry or flow virometry, respectively. The means and SD of at least three independent viral preparation for each viral isolate were determined. (e and f) Comparison of the binding efficiency of nnAbs and bNAbs on viral particles and producer cells. Raw data of the percentages of anti-Env antibodies binding on transfected cells (e) and on viral aggregates (f). Each dot corresponds to an individual independent staining. All bNAbs or nnAbs listed in Fig. 4 have been grouped together. Statistical analysis was performed using Prism software and the Mann-Whitney t test. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.