Figure 5: Epithelial HDAC3 regulates expression of the lipid transporter gene Cd36 by coactivating ERRα through PGC-1α.
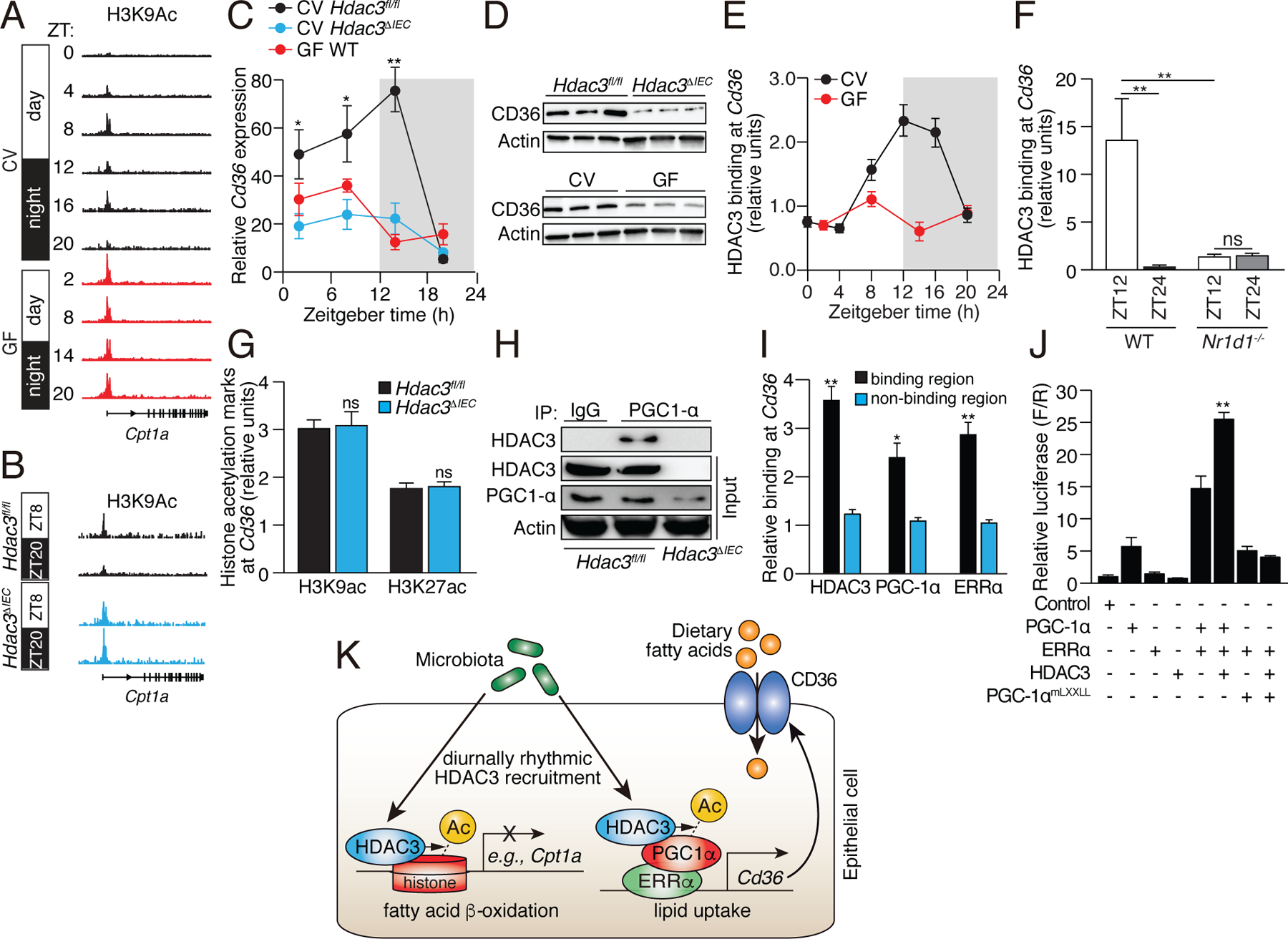
(A,B) Genome browser view of the Cpt1a locus, showing H3K9ac marks in small intestinal epithelial cells from CV and GF WT mice (A), and CV Hdac3fl/fl and Hdac3ΔIEC mice (B). Statistical test for circadian rhythms: P=0.0007 for CV, P=0.504 for GF, and P=0.00012 for Hdac3fl/fl and Hdac3ΔIEC mice. (C,D) Diurnal expression of Cd36 transcript (C) and protein (D) in IECs from Hdac3fl/fl CV mice, Hdac3ΔIEC CV mice and GF WT mice. N=3 mice per group. (E) ChIP-qPCR analysis of HDAC3 binding at the Cd36 promoter in CV and GF mice across a day-night cycle. (F) Relative abundance of bound HDAC3 at Cd36 in small intestinal epithelial cells from WT and Nr1d1−/− mice as determined by ChIP-qPCR analysis. N=3 mice per time point per group. (G) ChIP-qPCR analysis of H3K9ac and H3K27ac marks at Cd36 in IECs from Hdac3fl/fl and Hdac3ΔIEC mice (at ZT14). (H) Co-immunoprecipitation of endogenous HDAC3 and PGC-1α in IECs from Hdac3fl/fl and Hdac3ΔIEC mice. N=3 pooled biological replicates per lane. (I) ChIP-qPCR analysis of HDAC3, PGC-1α and ERRα shows co-localization at the Cd36 promoter in IECs from CV WT mice. “Binding region” refers to a known binding site for each protein at the Cd36 locus in adipose tissue (17). N=3 mice per group. (J) Luciferase reporter assay of transcription driven by a Cd36 promoter, demonstrating combinatorial effects of HDAC3, PGC-1α and ERRα. Empty vector was used in the control group. N=3 per group. (K) Model showing how the intestinal microbiota regulates diurnal rhythms in epithelial metabolic pathways through HDAC3. *P<0.05; **P<0.01; ns, not significant by two-tailed t-test. Means±SEM (error bars) are plotted. ZT, Zeitgeber time; CV, conventional; GF, germ-free.
