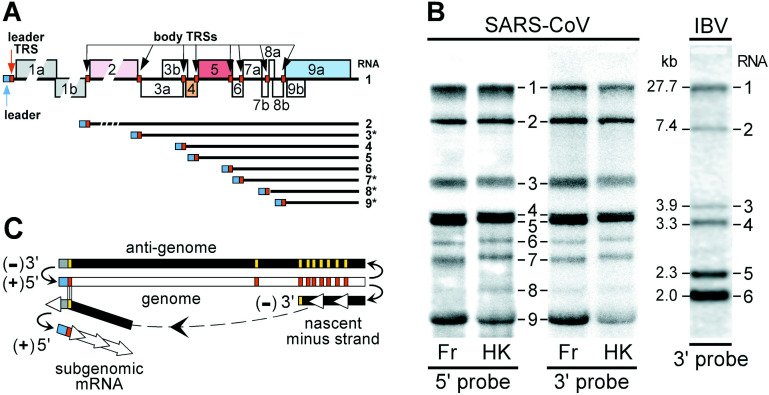
Figure 3.
SARS-CoV subgenomic mRNA synthesis. (A) Organization of ORFs in the 3′ end of the SARS-CoV genome with predicted leader and body TRSs indicated by small boxes. The subgenomic mRNAs resulting from the use of these TRSs for leader-to-body fusion are depicted below, with mRNAs predicted to be functionally bicistronic indicated with an asterisk (∗). (B) Hybridization analysis of intracellular viral RNA from Vero cells infected with SARS-CoV, Frankfurt-1 (Fr) and HKU-39849 (HK) isolates. See Materials and Methods for technical details. Oligonucleotides complementary to sequences from the SARS-CoV leader sequence and to a region in the genomic 3′ end both recognized a set of nine RNA species (the genome (RNA1) and eight subgenomic RNAs) confirming the presence of common 5′ and 3′ sequences. RNA from Vero cells infected with avian infectious bronchitis virus (IBV), which produces only five subgenomic mRNAs of known sizes41 was run in the same gel and used as a size marker. (C) Model for nidovirus subgenomic RNA synthesis by discontinuous extension of minus strands.19., 39. Whereas genome replication relies on continuous minus strand synthesis (antigenome), subgenomic minus strands would be produced by attenuation of nascent strand synthesis at a body TRS (red bar), followed by translocation of the nascent strand to the leader TRS in the genomic template. Following base-pairing between the body TRS complement at the 3′ end of the minus strand and the leader TRS, RNA synthesis would resume to complete the subgenomic minus strand that would then serve as template for the transcription of subgenomic mRNAs.