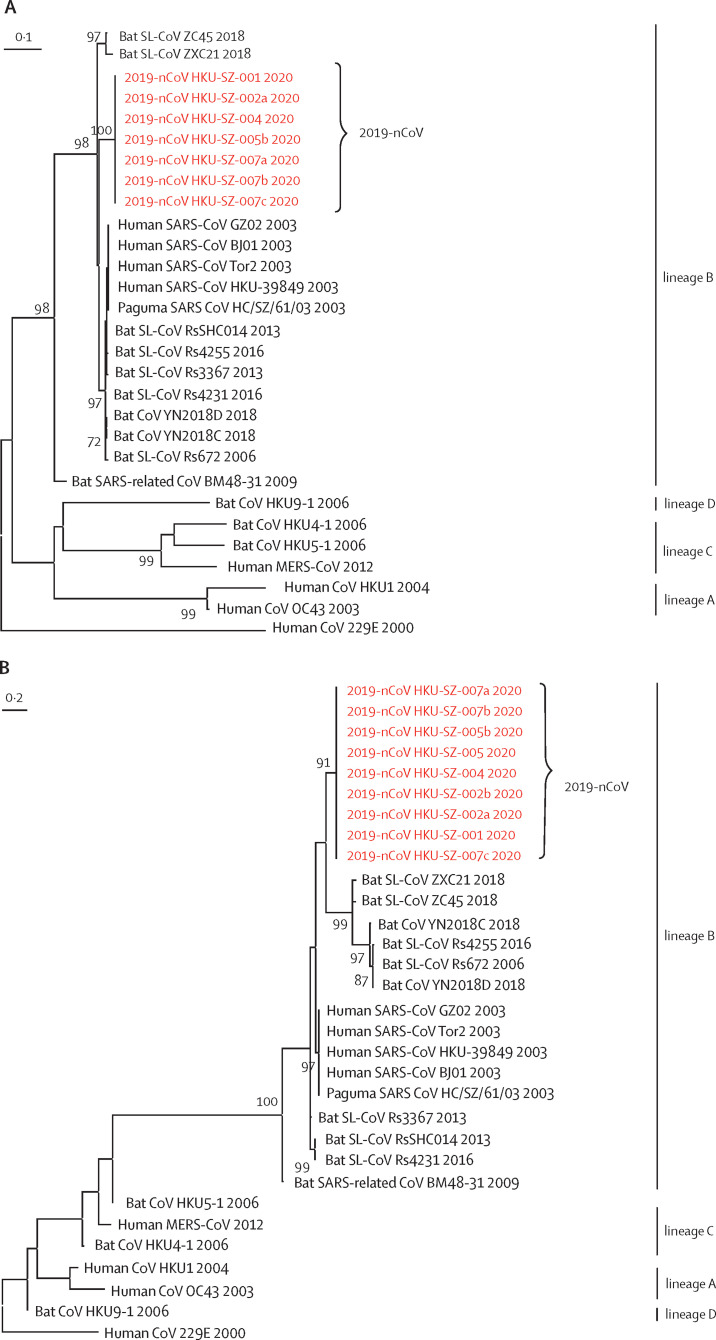
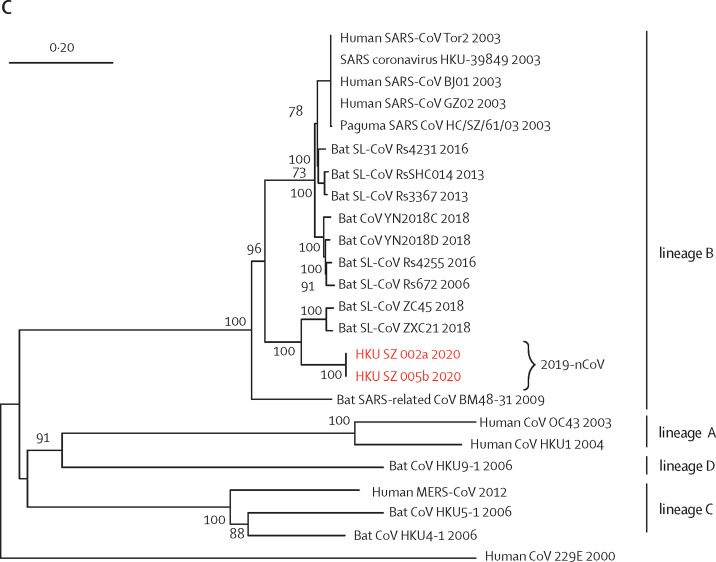
Figure 3.
Phylogenetic trees of genetic sequences
(A) Amplicon fragments of RNA-dependent RNA polymerase of patients 1, 2, 4, 5, and 7. (B) Amplicon fragments of Spike gene of patients 1, 2, 4, 5, and 7. (C) The full genome sequences of strains from patients 2 and 5. Red text indicates the coronavirus (CoV) strains detected in the patients in the present study. 2019-nCoV is 2019 novel coronavirus. HKU-SZ-001 refers to the strain detected in the nasopharyngeal swab of patient 1; HKU-SZ-002a refers to strain detected in the nasopharyngeal swab of patient 2; HKU-SZ-002b refers to strain detected in the serum sample of patient 2; HKU-SZ-004 refers to the strain detected in the nasopharyngeal swab of patient 4; HKU-SZ-005 refers to the strain detected in the throat swab of patient 5; HKU-SZ-005b refers to the strain detected in the sputum sample of patient 5; HKU-SZ-007a refers to the strain detected in the nasopharyngeal swab of patient 7; HKU-SZ-007b refers to the strain detected in the throat swab of patient 7; and HKU-SZ-007c refers to the strain detected in the sputum sample of patient 7 (appendix p 6). The NCBI GenBank accession numbers of the genome sequences are MN938384 (HKU-SZ-002a), MN975262 (HKU-SZ-005b), MG772934 (Bat SL-CoV ZXC21), MG772933 (Bat SL-CoV ZC45), AY274119 (hSARS-CoV Tor2), AY278491 (SARS coronavirus HKU-39849), AY278488 (hSARS-CoV BJ01), AY390556 (hSARS-CoV GZ02), AY515512 (Paguma SARS CoV HC/SZ/61/03), KY417146 (Bat SL-CoV Rs4231), KC881005 (Bat SL-CoV RsSHC014), KC881006 (Bat SL-CoV Rs3367), MK211377 (Bat CoV YN2018C), MK211378 (Bat CoV YN2018D), KY417149 (Bat SL-CoV Rs4255), FJ588686 (Bat SL-CoV Rs672), NC014470 (Bat SARS-related CoV BM48-31), EF065513 (Bat CoV HKU9-1), AY391777 (hCoV OC43), NC006577 (hCoV HKU1), NC019843 (hMERS CoV), NC009020 (Bat CoV HKU5-1), NC009019 (Bat CoV HKU4-1), and NC002645 (hCoV 229E).