Fig. 2. VP3 domain organization and structural comparisons.
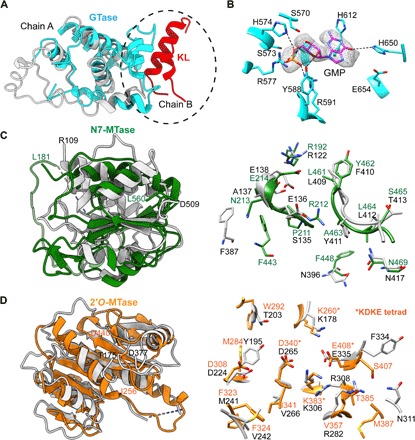
(A) Structural comparison of the GTase domains in RV VP3 (cyan) and BTV VP4 (light gray) showing similar structural folds. GTase domain of VP3 additionally participates in the interaction with the KL domain (helices 165 to 178, shown in red) from the neighboring subunit at the dimeric interface (circled). (B) Electron density for the GMP observed at the catalytic center of GTase domain in the x-ray structure. Conserved catalytic residues interacting with the GMP are denoted. (C) Structural comparison of the guanine N7-MTase (green) and (D) 2′-O-MTase (orange) domains in RV VP3 with those in BTV VP4 (light gray). Note that the substrate binding region shows ~50% conservation in amino acid sequences. The 2′-O-MTase domain also contains the well-characterized catalytic KDKE tetrad motif (in asterisks), which is a conserved feature of MTase domains of other capping enzymes. N- and C-terminal residues of the MTase domains in RV VP3 (L181 to L560 for N7-MTase and I256 to D440 for 2′-O-MTase) and BTV VP4 (R109 to D509 for N7-MTase and T175 to D377 for 2′-O-MTase) are indicated (domain colors as in previous figures).
