Abstract
Physical guidance from the underlying matrix is a key regulator of cancer invasion and metastasis. We explore the effects of surface topography on the migration phenotype of multiple breast cancer cell lines using aligned nanoscale ridges and asymmetric sawtooth structures. Both benign and metastatic breast cancer cells preferentially move parallel to nanoridges, with enhanced speeds compared to flat surfaces. In contrast, asymmetric sawtooth structures unidirectionally bias the movement of breast cancer cells in a cell-type dependent manner. Quantitative analysis shows that the level of bias in cell migration increases when cells move with higher speeds or with higher directional persistence. Live-cell imaging studies further reveal that actin polymerization waves are unidirectionally guided by the sawteeth in the same direction as the cell motion. High-resolution fluorescence imaging and scanning electron microscopy studies reveal that two breast cancer cell lines with opposite migrational profiles exhibit profoundly different cell cortical plasticity and focal adhesion patterns. These results suggest that the overall migration response of cancer cells to surface topography is directly related to the underlying cytoskeletal architectures and dynamics, which are regulated by both intrinsic and extrinsic factors.
Keywords: nanotopography, cell migration, actin dynamics, focal adhesion, breast cancer, metastasis, esotaxis
Graphical Abstract
The ability to migrate is conserved in a wide variety of cell types throughout the phylogeny and plays a pivotal role in many cellular processes.1–4 Cells typically undergo directional migration with a relatively high persistence5 rather than moving randomly.6 However, the biological and physical mechanisms governing directed cell migration in many physiological and pathological processes are still poorly understood. In addition to the intrinsic characteristics of cells, the extracellular environment plays an important role in the guidance of cell movement. Cells can be guided by single or multiple extracellular cues, including chemoattractant gradients (chemotaxis),7,8 stiffness gradients of the underlying substrate (durotaxis),9 electric fields (electrotaxis),10 and topographic structures on the surfaces (contact guidance).11,12
Cancer invasion and metastasis, processes in which active primary tumor cells migrate to adjacent tissues and eventually colonize distant organs, are major life-threatening events in cancer patients. Contact guidance is known to regulate cell adhesion, intracellular signaling, and directed migration13–15 and as such is a key component of cancer invasion. The role of the extracellular matrix (ECM) in guiding the three-dimensional (3D) migration of cancer cells in vivo has been studied extensively.14–18 For example, the directional migration of carcinoma cells can be enhanced by aligned collagen fibers, especially for breast cancer stem cell populations.17 Furthermore, Alexander et al. suggested that cancer cell dissemination patterns, including individual cell invasion, diffusively collective invasion, and strand-like invasion, are governed by the ECM structures that the tumor cells encounter.16 However, the topographic features dictating the invasion pattern remain unknown, as do the mechanisms by which tumor cells sense topographic features on a subcellular level. Nanofabrication techniques have inspired the rapid growth of materials that mimic natural in vivo topographies or 3D environments.19 Recent studies have shown that engineered nanotopographies can not only cause cells to recapitulate in vivo migrational behaviors20–22 but also influence various cell functions.23–25 Thus, nanopatterned substrates have the potential to provide insight into the physical and molecular mechanisms that regulate contact guidance during cancer progression.
The ability of patterned topographies to induce directional cell migration is a complex process that requires coordination between distinct cellular machineries, such as the assembly and disassembly of focal adhesions (FAs), the polymerization and depolymerization of actin filaments, and even the reorganization of organelles.26 The formation of a FA is initiated by the engagement of integrins with their corresponding ligands on the substrate, followed with the recruitment of focal adhesion kinase (FAK), paxillin, vinculin, and other associated proteins.27 The FA-associated proteins not only link the integrin receptors with the actin cytoskeleton28 but also regulate actin polymerization and contractility through Rho GTPase signaling.29 Furthermore, the actin cytoskeleton controls the protrusive activity of migrating cells, which in turn mediates the formation of adhesions. Recent studies from our group on Dictyostelium discoideum (D. discoideum) cells and neutrophils have shown that actin polymerization occurs preferentially near nanoridges, leading to guided traveling waves that can bias cellular movement bidirectionally.30 In the case of asymmetric nanotopographies this motion can be biased unidirectionally.31 It is also well established that various cell types elongate and exhibit aligned stress fibers when placed on ridges or grooves with widths and repeat distances on a subcellular scale.32–34 Ray et al. reported that the anisotropic force originating from local FAs and the concomitant alignment of actin fibers induces the polarization and migration of carcinoma cells along ridges, and that the guidance response varies considerably among different types of carcinoma cells.22 However, the link between the intrinsic heterogeneity of cancer cell lines and its influence on FA and actin dynamics during contact guidance is poorly understood.
Here, we study the migrational phenotypes of multiple breast cancer cell lines plated on aligned ridges and asymmetric sawtooth architectures. Data from long-term live-cell imaging enable us to perform extensive analysis of cell migration, actin polymerization dynamics, and FA distributions. Our findings elucidate the connection between the overall cell motility phenotype of breast cancer cells and their intracellular scaffolding architectures, adhesion complexes, and actin cytoskeleton and also provide insights into the biophysical mechanisms underlying contact guidance during cancer metastasis.
RESULTS AND DISCUSSION
Nanoridges Provide Bidirectional Guidance for Both Benign and Metastatic Breast Cancer Cells.
We compare the migration phenotypes of two cell lines belonging to the MCF10A cell series, a breast cancer progression model,35–37 to explore contact guidance in cell lines sharing a similar genetic background but with distinct metastatic abilities. MCF10A (also referred as M1), a benign epithelial human cell line, and its metastatic mutant cell line counterpart MCF10CA1 (also referred as M4) were studied on flat surfaces and on nanoridges composed of the same material. The M1 and M4 cells are negative for receptors for estrogen, progesterone, and HER2 (i.e., triple negative).38 Compared to immortalized epithelial M1 cells, the invasive M4 cells form tumors in the lungs of immunocompromised mice after tail vein injection.37,39
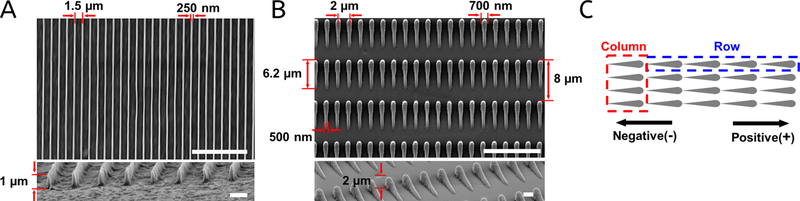
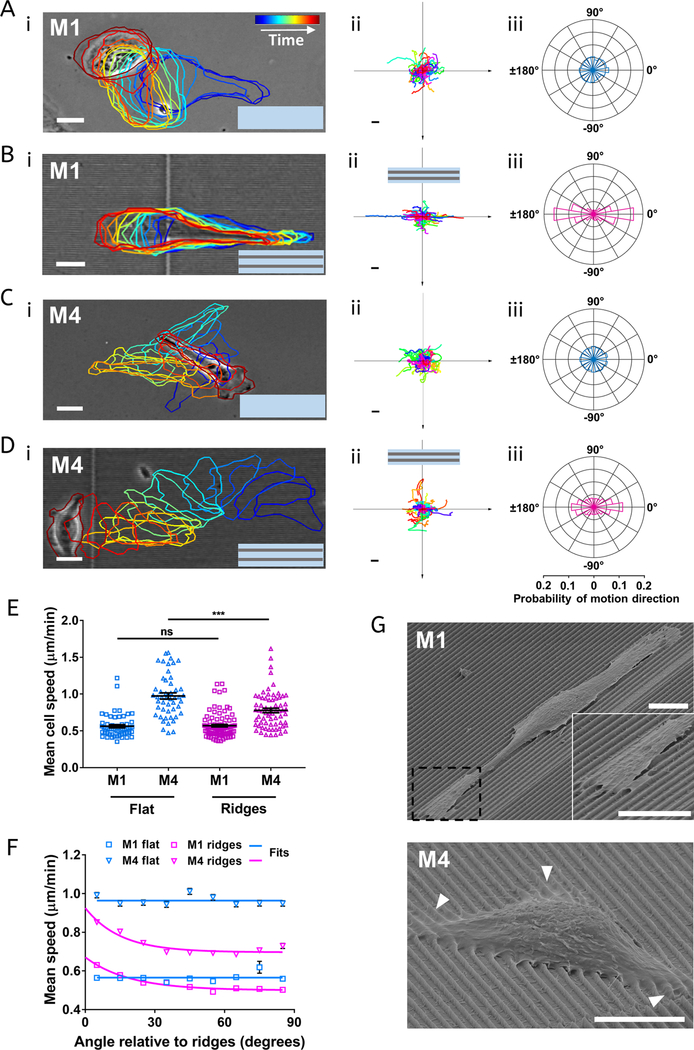
Arrays of parallel nanoridges or nanogrooves (where nano implies that their widths are on the nanoscale) have been used in previous contact guidance studies.20,40,41 Here, we use nanotopographies composed of an acrylic resin.42 Each nanoridge has a width of ∼250 nm and a height of ∼1 μm, and the spacing between the centers of adjacent nanoridges is 1.5 μm (Figure 1A). By tracing cell boundaries over time and generating spider plots of the cell tracks, we found that both M1 and M4 cells move in a random manner on a flat acrylic surface (Figure 2Ai,ii,Ci,ii). Polar histograms of the directions of both M1 and M4 cells demonstrate that there is no preferred direction of motion, as expected for a microenvironment in which no chemoattractant gradient nor other guidance cue is present (Figure 2Aiii,Ciii). However, M1 and M4 cells migrating on nanoridges move preferentially parallel to the nanoridges (Figure 2Bi,ii,Di,ii), as is also seen in polar histograms (Figure 2Biii,Diii). This finding is consistent with previous results showing that parallel micro- or nanoscale grooves and ridges induce bidirectional guidance in a variety of cell types.30,40,41,43,44 M1 cells exhibit a notably stronger guidance response to the nanoridges than do M4 cells (Figure 2Biii,Diii), suggesting that the ability of cells to follow guidance cues is cell-type-dependent. We also found that, although the nanoridges provide guidance for cell migration, they do not increase the mean cell speed compared to the flat resin surface and, in the case of M4 cells, actually decrease the mean speed (Figure 2E). To explore this behavior in more detail, we measured the mean value of all velocities within a given angle range relative to the orientation of the nanoridges. We define the angle of motion as ranging from −180 to +180°, with the ridge direction along the x axis. Because the ridges are aligned horizontally in our images of ridges, on flat surfaces, the horizontal direction in images was also defined as the x axis. We found that, on flat surfaces, the mean velocity value is independent of angle for both cell types, within our experimental uncertainty (Figure 2F). However, on nanoridges, both M1 and M4 cells exhibit a mean velocity when migrating along the ridge orientation higher than that when migrating perpendicular to it (Figure 2F). To confirm the significance of this increasing trend, we fitted the experimental results with a horizontal line model and a one-phase decay model and compared the robustness of two models for each curve. We found that for M1 and M4 cells on flat surfaces, the fit of the horizontal line model is preferred; however, the fit of the one-phase decay model is preferred for the two cell lines on nanoridges (P < 0.0001 and P = 0.001 for M1 and M4 cells, respectively). Together, these findings show that cells not only migrate preferentially along nanoridges but also migrate faster along them than perpendicular to them.
Figure 1.
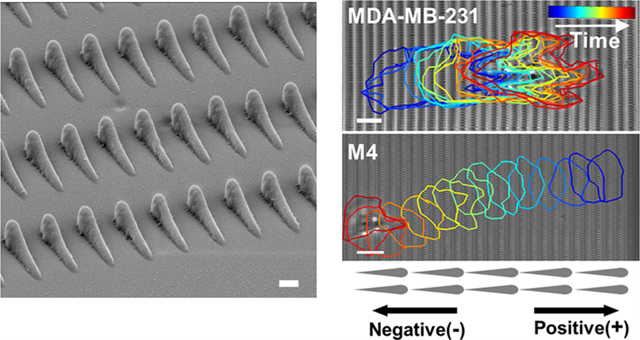
Nanotopographies used in this study. Top-view and side-view SEM images of (A) nanoridges and (B) asymmetric sawteeth used in this study. The scale bars in the top- and side-view images are 10 and 1 μm, respectively. (C) Schematic depicting a sawtooth row (blue box) and a sawtooth column (red box), as well as the direction defined as positive.
Figure 2.
Nanoridges provide bidirectional guidance for benign and metastatic breast cancer cells. (A−Di) Bright-field images of M1 or M4 cells on a flat surface (A,C) and on nanoridges (B,D). The colored outlines depict how the cell morphology changes over time, which increases from blue to red. The scale bar is 10 μm. (A−Dii) Centroid motion tracks of M1 and M4 cells, respectively, on a flat surface (A,C) and on nanoridges (B,D); n = 56 tracks (Aii); 94 tracks (Bii); 49 tracks (Cii); 68 tracks (Dii). The scale bar is 60 μm. (A−Diii) Probability distributions of cell motion directionality. The scale at the bottom corresponds to the horizontal axis in each rose plot. (E) Average cell speed of M1 and M4 cells on flat surfaces and on nanoridges. The middle solid line in each column is the mean value, and the bar is standard error of the mean; ***p < 0.001 (unpaired t test). (F) Average instantaneous speed of M1 and M4 cells relative to the orientation of ridges (0° along ridges, 90° perpendicular to ridges). Squares and triangles are experimental results, and solid lines are the fits to the experimental results. Blue solid line was fitted to a horizontal line, and the pink solid line was fitted to a one-phase decay model. (G) Representative SEM images of M1 (top) and M4 (bottom) cells on nanoridges. The magnified image highlights the anchoring of M1 cell lamellipodia on top of the nanoridges. White arrows indicate sites where M4 cell lamellipodia curve over the nanoridges. The scale bar is 10 μm.
Scanning electron microscopy (SEM) provides further insights into how cells interact with the nanoridges. On nanoridges, the lamellipodia of M1 cells form along nanoridges on both sides of the cell and are anchored on top of the nanoridges, without much penetration into the regions between nanoridges (Figure 2G). One possible explanation for this phenomenon is that the nanoridges guide M1 cells bidirectionally because lamellipodium formation is promoted along the ridge direction and/or is suppressed perpendicular to the ridge direction. In contrast, M4 cells form lamellipodia that curve over nanoridges and into the bottom of the regions between nanoridges, and typically span several nanoridges (Figure 2G, white arrows). Because contact guidance relies upon the spatial control of cell protrusions,21 the behavior of the lamellipodia of M4 cells likely contributes to their lower contact guidance as compared to M1 cells.
Asymmetric Sawteeth Bias the Movement of Breast Cancer Cells in a Cell-Type-Dependent Manner.
We next consider the ability of breast cancer cells to respond to asymmetries in texture using sawtooth-shaped asymmetric nanotopographies. Each sawtooth has a maximum height of ∼2 μm, a length of ∼6.2 μm, and a width of ∼700 nm at the head and ∼500 nm in the center (Figure 1B). The height of an individual sawtooth increases gradually from its tail to its head and then drops rapidly. The pitch of the sawteeth is 8 μm along the sawtooth direction and 2 μm perpendicular to the sawtooth direction. We define a line of sawteeth along the sawtooth direction as a row. As all of the rows are in registry, we define a line of sawteeth perpendicular to the rows as a column (Figure 1C). We define the direction from the tail of a sawtooth to its head (up the slope) as the positive direction along the x axis.
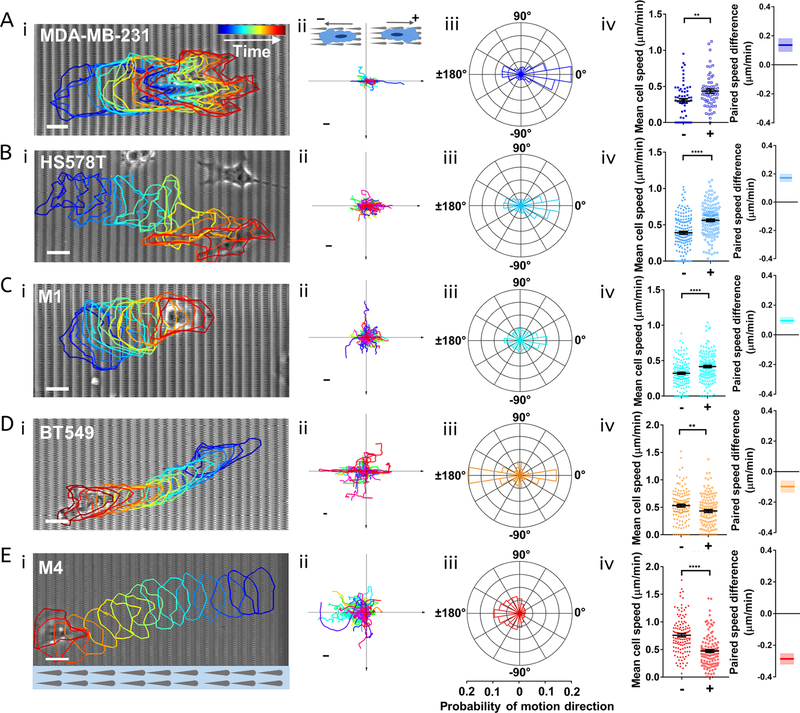
We found that M1 cells plated on the sawteeth move preferentially in the positive direction with an anterior lamellipodium (Figure 3Ci,ii). A polar histogram of directionalities shows a higher percentage of orientation angles around 0 than 180° (Figure 3Ciii). In contrast, M4 cells exhibit amoeboid-like migration, with ellipsoidal cell shapes and multiple pseudopods (Figure 3Ei).45,46 Furthermore, the majority of the M4 cell tracks face in the negative direction (Figure 3Eii), as do the cell velocities (Figure 3Eiii). The preference of the M4 cells to move in the negative direction is considerably stronger than that of the M1 cells to move in the positive direction. Unlike M1 cells, M4 cells are more likely to move perpendicular to the sawteeth than to move in the positive direction.
Figure 3.
Asymmetric sawteeth bias the movement of various breast cancer cell lines. (A−Ei) Bright-field images of (A) MDA-MB-231, (B) HS578T, (C) M1, (D) BT549, and (E) M4 cells migrating on sawteeth. The colored outlines illustrate how the cell morphology changes over time, which increases from blue to red. The scale bar is 10 μm. (A−Eii) Centroid motion tracks of (A) MDA-MB-231, 62 tracks, (B) HS578T, 151 tracks, (C) M1, 143 tracks, (D) BT549, 126 tracks, and (E) M4, 122 tracks cells on sawteeth. The scale bar is 60 μm. (A−Eiii) Probability distributions of cell motion directionality. The scale at the bottom corresponds to the horizontal axis in each rose plot. (A−Eiv) Average cell velocity components of the cell lines along the positive direction (+) and the negative direction (−) and paired speed difference for each cell. The middle solid line is the mean value, and the bar is standard error of the mean. The shaded areas represent standard errors in paired speed difference plots; *p < 0.05, **p < 0.01, ****p < 0.0001.
We also measured the behavior of three other triple-negative human breast cancer cell lines, HS578T, MDA-MB-231, and BT549. HS578T and BT549 are carcinoma cell lines derived from primary tumors. MDA-MB-231 is a metastatic carcinoma cell line from pleural effusion.47 We found that MDA-MB-231 and HS578T cells exhibit a mesenchymal migration mode on the sawteeth, with multiple competing protrusions (Figure 3Ai,Bi).46,48 Spider plots of cell tracks and polar histograms indicate that these cells tend to move in the positive direction (Figure 3Aii,iii,Bii,iii). In contrast, BT549 cells migrate in a blebby (and slightly elongated) manner on the sawteeth (Figure 3Di). The BT549 cells preferentially move along the sawteeth, with a somewhat greater probability of moving in the negative direction than in the positive direction (Figure 3Dii,iii). We also found some motion of BT549 cells perpendicular to the sawteeth (Figure 3Diii), as with the M4 cells. To assess the behavior of the cells on the sawteeth further, we grouped the velocity component along the sawtooth axis into motion in the positive and negative directions, + and −, respectively. The velocities in each group were then averaged separately for each cell, and a paired speed difference for each cell was determined. We found that MDA-MB-231, HS578T, and M1 cells have higher mean speeds in the positive direction than in the negative direction, but the difference is smaller for M1 cells than for the other two cell lines (Figure 3Aiv,Biv,Civ). For the BT549 and M4 cells, the mean speeds in the positive direction are lower than those in the negative direction (Figure 3Div,Eiv). BT549 cells exhibit less paired speed difference and less unidirectional contact guidance compared with M4 cells. Together, these results show that the sawteeth provide both contact guidance and unidirectional bias in migration and speed for the breast cancer cell lines studied here. Both of these phenotypes are dependent on the cell type.
We previously reported that D. discoideum cells and neutrophils exhibit biased directional migration when plated on sawtooth surfaces.31 This unidirectional topographic guidance, which is achieved by an asymmetric textures on a subcellular scale, is termed microthigmotaxis.31 Topographic gradients represent another way to guide cell migration unidirectionally–a phenomenon called topotaxis.49 Kim et al. reported that individual fibroblasts are able to sense the anisotropic gradient of ridged patterns and modify their morphology and migration in response to the local pattern density.32 On a 2D gradient lattice patterned substrate, the fibroblasts preferentially migrated toward the denser area and ultimately accumulated at the denser zones.50 Anisotropic variation of the density of nanoposts along one direction has also been reported to lead to the biased migration of melanoma cells.51 Recent work from Comelles et al. showed that large asymmetric ratchet-like structures (at the scale of single cells) biased the motion of fibroblasts toward the positive direction of the ratchet pattern, and that this ratchet guidance was improved by superimposing a fibronectin gradient.52 Interestingly, even without the topographic ratchet pattern, aligned ratchet-shaped adhesive fibronectin patches were able to induce a biased long-term motion that was determined by asymmetric protrusion formation.53,54 First introduced by the physicist Richard Feynman, this type of guidance is termed ratchetaxis.55 In contrast with ratchet-like topography, the asymmetry induced by the nanoscale sawteeth in our study is at the subcellular level, suggesting that directed migration can stem from the accumulation of small biases in intracellular elements. Furthermore, we show that breast cancer cells exhibit microthigmotactic behavior that depends strongly on cell type. Although the underlying mechanism remains to be determined, it is likely that the distinct mutation patterns and intrinsic heterogeneities of the different cell lines are reflected in the cells’ contact guidance and directional bias behaviors. We speculate that such significant differences may be related to the metastatic potential of the different cancer cell lines in vivo.
Degree of Bias in Cell Migration Is Related to Cell Speed and Long-Term Persistence.
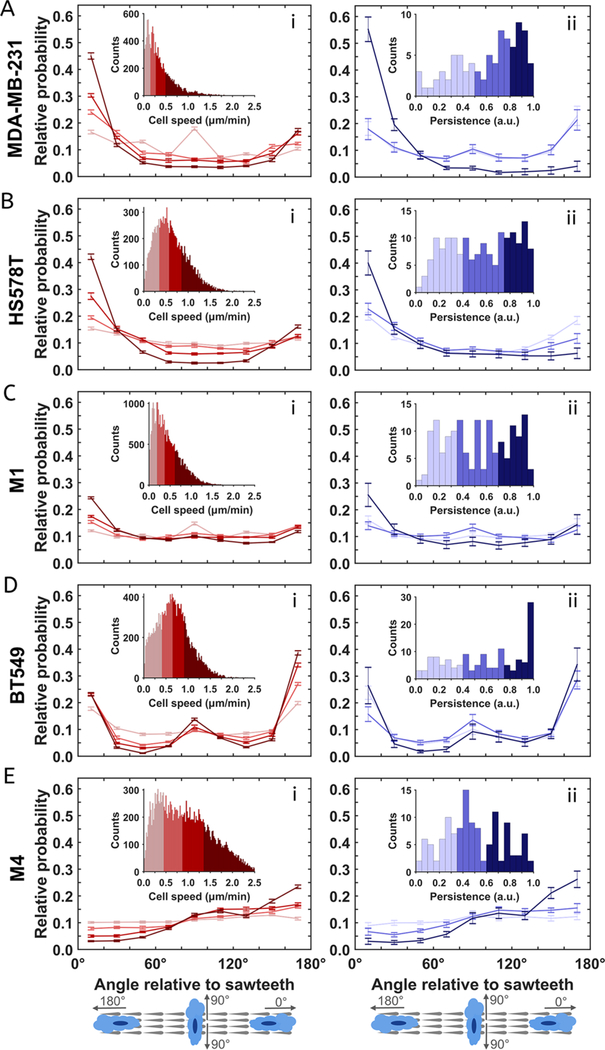
Cell migration is a heterogeneous process, in which different cells move with distinct velocity and persistence. Indeed, the direction and speed of any given cell also vary over time. To study how cell speeds influence unidirectional guidance on sawteeth, we divided histograms of the cell speeds into four quartiles (see insets of Figure 4Ai–Ei). We also plotted the normalized probabilities of the angle of cell motion relative to the sawtooth positive direction for each quartile (Figure 4Ai–Ei). For HS578T and MDA-MB-231 cells, there is no observable directional preference at low speeds, but migrational bias increases with increasing cell speed (Figure 4A,Bi). This trend is similarly robust for these two cell lines. In contrast, the level of directional preference of M1 cells remains low even at higher speeds, suggesting that speed and contact guidance are not strongly coupled in these cells (Figure 4Ci). BT549 and M4 cells behave like HS578T and MDA-MB-231 cells: unidirectional guidance increases with increasing speed (Figure 4Di,Ei). The trend is less evident for BT549 cells because the probability curves are considerably overlapping. Nevertheless, the probability of BT549 cells migrating along the sawtooth negative direction (∼180°) keeps increasing with speed, whereas the probability for migration along intermediate angles decreases.
Figure 4.
Level of biased cell motion increases with cell speed and long-term persistence. Column (i) shows the probability of the direction of motion based on speed for (A) HS578T, (B) MDAMB-231, (C) M1, (D) BT549, and (E) M4 cells. The insets show the speed distributions of the cell lines and the ranges of four sections which were equally divided based on the total counts of cell speeds. The cell speed increases from light red to dark red in both the plots and the insets. Column (ii) shows the probability of the direction of motion based on migrational persistence for the same cell lines. The insets show the long-term persistence distributions of the cell lines and the ranges of tertiles based on the total counts of cells. The long-term persistence increases from light blue to dark blue in both the plots and the insets. All error bars represent the standard error of the mean.
Using a similar segmentation approach, we investigated the effect of long-term persistence of cell motion on the unidirectional guidance of migration. We define the long-term persistence of cell motion as the ratio of the total displacement of the cell to the total length of the cell track for a chosen time interval. A cell moving in a straight line has a long-term persistence of 1, whereas random motion leads to a longterm persistence of 0. Cells were separated into three equal fractions based on their distributions of cell persistence (see insets of Figure 4Aii–Eii). The normalized probabilities of the cell motion direction were then calculated by using all velocities from the cells in each of the three fractions. We found that cells with the greatest long-term persistence show the strongest contact guidance and bias toward the preferred direction of each cell line (Figure 4Aii–Eii). However, the directional preference is similarly vague for cells with low and intermediate long-term persistence across all cell lines (Figure 4Aii–Eii). For BT549 cells, the probability curves from all three fractions greatly overlap within the estimated uncertainty, suggesting that the correlation between contact guidance and cell persistence is weak in these cells. Together, we conclude that the propensity of cells to migrate in a biased fashion increases when the cells move with higher speeds or with higher directional persistence. Recently, a universal coupling between cell speed and cell persistence (UCSP) was discovered and proved to be valid across a variety of cells and dimensional settings.56 To test whether this UCSP law can be applied to our system, we plotted the persistence time versus mean instantaneous speed for all cell lines by following the definition of persistence time and the quantification method in Maiuri et al.56 Interestingly, we also found a correlation between the persistence time and mean instantaneous speed at low speeds and a rapid saturation at high speeds for all breast cancer cell lines, except for M4 cells, where we observed a second rising phase at high speeds (Figure S1). We speculate that the rapid saturation is due to following reasons. First, in our experimental setup, the cell density is higher and cell trajectories are not long compared with the plating conditions in Maiuri et al. Second, the UCSP law assumes a linear coupling between cell speed and actin flow, which may not be valid in our setup due to the complexity induced by the sawtooth structures. Nevertheless, it will be interesting to further examine whether the UCSP law holds for topography-guided migration.
Unidirectional Actin Polymerization Drives Biased Cell Migration.
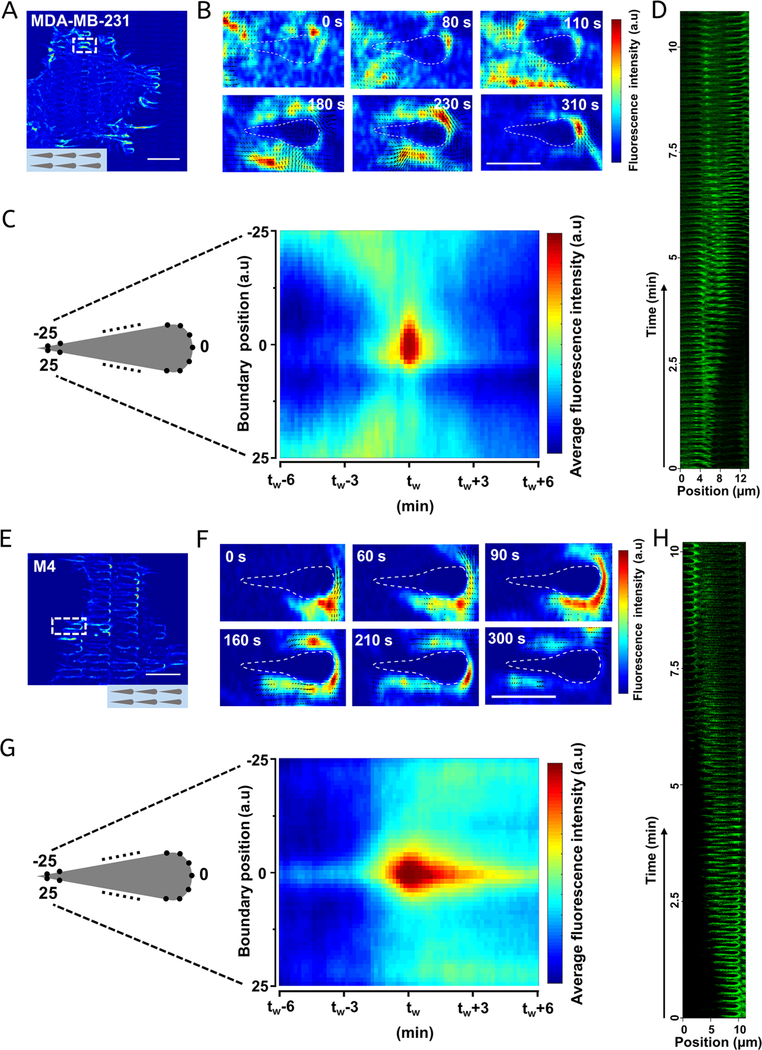
During migration and metastasis, each cancer cell dynamically reorganizes its cytoskeleton, including actin structures and microtubules, to maneuver and to adapt to complex and changing environments. Filopodia and lamellipodia are actin-rich structures that act as dynamic sensors with which cells detect their surrounding microenvironment.21,40,57 The formation of filopodia and lamellipodia requires extensive rearrangement of cytoskeletal elements. Driscoll et al. found that actin polymerization in D. discoideum migrating on nanoridges is nucleated near the nanoridges, after which actin polymerization proceeds in a wave-like manner along the ridges (a process called esotaxis).30 Further, it has been established previously that the unidirectionally guided motion of D. discoideum cells on lines of sawteeth (microthigotaxis) arises from esotaxis, which shares the same direction as cell motion.31 We therefore explored the correlation between the directional bias in actin polymerization and cell motion for the MDA-MB-231 and M4 cell lines, as they represent two opposite extremes of contact guidance and bias. To assess actin dynamics, we used cells infected with a fluorescence-tagged protein (Lifeact-eGFP), which associates with polymerizing F-actin. Time-lapse recordings of Lifeact-eGFP/MDA-MB-231 cells reveal that these cells preferentially form distinct F-actin structures surrounding the nanosawteeth at the leading edges of cell protrusions (Movie S1). Actin polymerization bursts occur occasionally in the middle of migrating cells and diffuse across the nanosawteeth (Movie S1). We used an optical-flow algorithm that will be described in detail elsewhere (Lee et al., unpublished results) to examine the local direction of the actin polymerization flux. Zooming in on one representative sawtooth (Figure 5A,B), we see that actin polymerization was initiated at the tail of the sawtooth and propagated toward the head on both sides of the sawtooth in a wave-like manner. The black arrows in Figure 5B represent optical-flow vectors, which are related to the local directionality of the actin waves. The optical-flow analysis indicates that actin polymerization is nucleated at the back of the sawtooth, after which the polymerization front moves forward on both sides of the sawtooth. Finally, the waves terminate at the front of the sawtooth (Figure 5B and Movie S2), consistent with our previous findings in D. discoideum.31
Figure 5.
Sawteeth trigger unidirectional actin polymerization in the same direction as the cell motion in a cell-type-dependent manner. (A,E) Fluorescence images of Lifeact-eGFP/MDA-MB-231 (A) or Lifeact-eGFP/M4 (E) cells on sawteeth. The colors indicate the intensity of fluorescence. The scale bar is 20 μm. (B,F) Actin flux at different time points around a single sawtooth highlighted by the white box in (A) and (E), respectively. The boundary of the sawtooth was generated based on the corresponding bright-field image and, due to diffractive effects, does not correspond to the actual shape of the sawtooth. The different colors represent different fluorescence intensities. The black arrows illustrate the direction of actin polymerization flux. The scale bar is 5 μm. (C,G) Averaged kymograph of actin intensity along one sawtooth boundary over time for (C) Lifeact-eGFP/MDA-MB-231 and (G) Lifeact-eGFP/M4 cells. The x axis is the relative time difference from defined time point, tw, when the actin polymerization wave initiates or reaches the sawtooth position 0. The length of the x axis is 71 frames (11.83 min). The y axis is the scaled position along one sawtooth boundary, for which 0 represents the front and ±25 represents two symmetric points near the tail. The different colors represent different fluorescence intensities. (D,H) Space/time plots of actin polymerization along two adjacent sawteeth in a sawtooth row for (D) Lifeact-eGFP/MDA-MB-231 and (H) Lifeact-eGFP/M4 cells.
To assess the average actin flow around a sawtooth, we extracted the boundary of every sawtooth from the bright-field images, measured the average intensity at each of the 51 boundary points of each sawtooth, and generated a kymograph representing all boundary points (on the y axis, −25 to +25) over time (on the x axis). The average kymograph for all of the sawteeth shows that the average fluorescence intensity is higher at the tails of the sawteeth (position ±25) at time tw − 6, suggesting that the initiation of actin polymerization occurs there. After time tw − 6, the average actin fluorescence slowly increases at the sides of the sawteeth and gradually peaks at position 0 around time tw. These findings support the conclusion that actin polymerization is nucleated at the tails of the sawteeth and moves toward the heads (Figure 5C). A space/time plot also shows that the actin waves propagate from one sawtooth to the adjacent one in the positive direction with a speed of ∼1 μm/min (Figure 5D). Thus, the unidirectional guidance of actin polymerization is in the same direction as that of the cell motion.
In Lifeact-eGFP/M4 cells, actin polymerization structures form not only at the cell protrusions but also in the middle of each cell. The actin polymerization intensity is always higher at the front of the sawteeth (Figure 5E and Movie S3). Data from a representative sawtooth reveal that actin polymerization is nucleated at the head of the sawtooth, after which the wavefront flows toward the tail of the sawtooth (Figure 5F and Movie S4). The average kymograph of actin polymerization activity around the sawteeth confirms that actin polymerization is nucleated at the heads of sawteeth and flows toward the tails along both sides (Figure 5G). The average kymograph also shows that the sawteeth heads maintain a relatively higher level of actin fluorescence intensity compared to other locations on the sawteeth after time tw, as illustrated in Movie S3. This may indicate that the curvature of the sawteeth heads promotes actin polymerization. The actin waves also are observed to travel from one sawtooth to the adjacent one in the negative direction (Figure 5H).
Microtubules are another essential cytoskeletal component of cells. Microtubules have been shown to be the first cytoskeletal element to be aligned by micron-scale grooves during the spreading of fibroblasts.58 This process is followed by the alignment of focal contacts, actin filaments, and finally cells.58 Microtubules were found to align with ridges on ridged substrates.42 Because microtubules regulate FA disassembly through integrin recycling and trafficking,59,60 it is natural to consider that microtubules play an important role in cell polarization and migration during contact guidance. To investigate this idea, we fixed MDA-MB-231 and M4 cells migrating on sawteeth and stained for microtubules using an antibody against tubulin. We found that the microtubules of MDA-MB-231 cells align with the sawtooth patterns. This alignment is particularly evident at the cell protrusions, where microtubules coil around the sawteeth (Figure S2A, white arrows). This phenomenon was not observed in M4 cells (Figure S2B), suggesting that the mechanisms that regulate microtubule dynamics differ in cells that respond differently to a specific nanotopographic surface.
These findings show, as previously observed in D. discoideum and neutrophils,31 that there is a direct connection between the migrational response to sawteeth and the underlying cytoskeletal dynamics. In other words, the esotactic guidance of the actin cytoskeleton plays an important role in microthigmotaxis. Actin waves are known to be important in cell membrane dynamics and motility. It has previously been shown that actin polymerization drives the wave-like movement of its upstream nucleator through an autoinhibitory mechanism,61 which can further generate the directional motion of cells.62,63 By using asymmetric sawtooth surfaces, we expand this concept by showing that an asymmetric topography can induce esotactic (actin) and microthigmotactic (migrational) behaviors in a cell-type-dependent manner. This dependence of the direction of these phenomena on cell type suggests that multiple regulatory nodes contribute to the fine control of actin dynamics. Some of these nodes may be intrinsic, including the antagonistic interplay between GTPase signaling and the activation of different actin nucleators,61,64 whereas others may be extrinsic, such as the use of integrin based complexes to interact with the underlying topographic guidance cues.
Distinct FA Patterns and Cortical Plasticity Promote Biased Cell Migration.
In addition to the actin and microtubule cytoskeletal networks, FAs are key component in contact guidance. FAs transduce extracellular forces from the ECM and induce a wide range of biochemical signals that go on to regulate cell adhesion and motility.29 In this context, Ohara and Buck first proposed that the polarization and alignment of cells on ridges results from the need to maximize the area of focal contacts.65 Several studies have now suggested that FA formation represents one plausible mechanism by which contact guidance is regulated.22,33,34,66,67 We examined the distribution pattern of FAs for both MDA-MB-231 and M4 cells on sawteeth using high-resolution fluorescence imaging of actin and paxillin, the latter of which is an important scaffold protein of FAs that recruits and integrates numerous structural and signaling proteins for the transduction of ECM signals.68
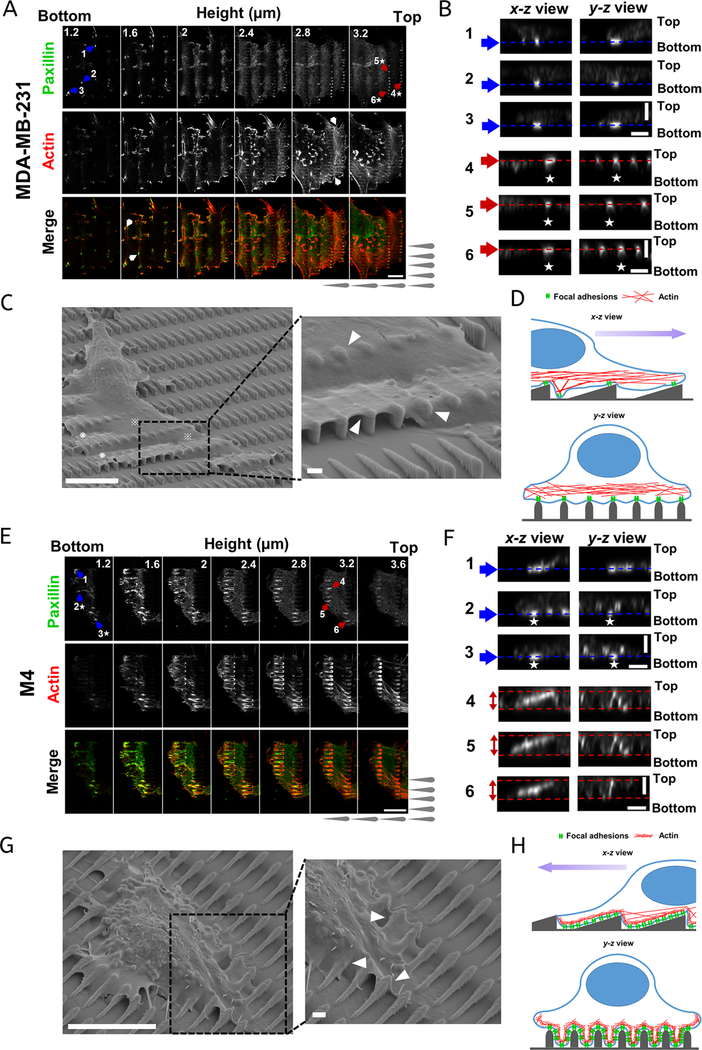
To capture the paxillin fluorescent signal around each sawtooth on the z axis (i.e., along the surface normal), we imaged cells at z positions ranging from below the bottom of the surface to above the top of each sawtooth, resulting in each sawtooth falling into a range of 1.2 to 3.2 μm on the z axis. A montage of paxillin and/or actin labeling of a MDA-MB-231 cell is presented in Figure 6A. The paxillin signal is first observed at a z of approximately 1.2 μm, which is at the bottom of the cell. The majority of the FAs are located around the cell periphery, at the tips of filopodia (Figure 6A; paxillin and merge channels). FA complexes are also seen in the gaps between adjacent sawtooth columns that connect with stress fibers (Figure 6A; see white arrows in merge channel and Movie S5). Above z = 2.0 μm, no obvious FAs are observed. The actin filament network starts to show up in cell lamellipodia and becomes obvious at z = 2.8 μm (Figure 6A; see white arrow in actin channel). Surprisingly, above z = 2.8 μm, the FA punctae are well organized, with a spacing similar to that of adjacent rows of sawteeth (Figure 6A; paxillin channel). To show precisely the spatial positions of the FAs in different z slices, we selected FA complexes from either the bottom layer (Figure 6A,B; 1−3, blue arrows) or the tips of the sawteeth (Figure 6A,B; 4−6, red arrows) and generated x−z and y−z projections (Figure 6B). The FAs from the bottom layer reach a peak intensity at a z of approximately 1.3 μm (Figure 6B; blue dashed lines and Figure S3B; top panel). In contrast, the ordered FA complexes from the top layer are located near the tips of the sawteeth and exhibit a maximum intensity at a z of approximately 2.9 μm (Figure 6B; red dashed lines and Figure S3B; bottom panel). Notably, strong fluorescence signals are not observed in the gaps between two adjacent FAs on the top of the sawteeth, suggesting that there is little membrane penetration into the grooves of the structures (Figure 6B; 4−6 y−z view). Because FA complexes are indicators of the cell-ECM interfaces, we speculate that the MDA-MB-231 cells primarily interact with the tips of the nanosawteeth and that the FAs and stress fibers sustain most of the cortex above the grooves.
Figure 6.
MDA-MB-231 and M4 cells exhibit distinct cortical plasticity and focal adhesion patterns. (A,E) Fluorescence image montage of paxillin (top), actin (middle), and the merge of the two (bottom) at different z positions for (A) MDA-MB-231and (E) M4 cells. Blue and red arrows highlight the focal adhesions chosen for further analysis in (B) and (F). The white arrows in the actin channel of (A) highlight the actin filament network. The white arrows in merge channel of (A) indicate focal adhesions located between two sawtooth columns. The scale bar is 10 μm. (B,F) The x−z and y−z views of representative FAs from the lowest and highest z positions in (A) and (E), respectively. The white stars indicate the representative FAs in the images in the x−z and y−z views. The single red and blue dashed lines in (B) mark the center z positions of the chosen FAs and the double red dashed lines in (F) indicate the expansion ranges of the chosen FAs. The scale bar is 2 μm. (C,G) Representative SEM images of (C) MDA-MB-231 and (G) M4 cells migrating on sawteeth. Magnified images of the areas are highlighted by the boxes with black dashed lines. White arrows point out the places showing distinct cell cortical plasticity of MDA-MB-231 and M4 (G). The scale bars are 10 μm in the zoomed-out images and 1 μm in the magnified images. (D,H) Schematic representation of the biased contact guidance for (D) MDA-MB-231 and (H) M4 cells on sawteeth. The purple arrow represents the direction of motion.
Indeed, SEM images of MDA-MB-231 cells on sawteeth show that the whole cell body remains on top of sawteeth like a “floating rigid blanket” (Figure 6C). The leading lamellipodia spread across multiple sawteeth in the same column, whereas the membrane rests on the top of the sawteeth with no penetration into the grooves between rows of sawteeth. This behavior is further highlighted by zooming in at the front of the cell lamellipodium (Figure 6C; zoomed-in image). This bridging behavior is in agreement with our results on M1 cells on nanoridges (Figure 2G; top panel), as well as with many previous reports.20,34,65,67 Bridging is thought to promote cellular alignment and anisotropic traction forces because of confined FA growth.22 In fact, this bridging effect may provide an explanation for our previous observation that M1 cells preferentially form FAs on ridges.42 At the front of a sawtooth, the cell membrane curves somewhat and wraps around the head (Figure 6C; white arrow heads). M1 cells, whose migration bias is in the same direction as that of MDA-MB-231 cells, exhibit a similar phenotype (Figure S3A). When the cell lamellipodia extend from one sawtooth column to another, the membrane can either bridge the gap (Figure 6C; see crosshatch symbol) or droop down to the bottom surface (Figure 6C; see asterisk). We envision that cell areas where the cell interacts with the bottom ECM (marked by ∗) provide support for the cell body, as observed by fluorescence imaging of actin and paxillin (Figure 6A).
Profoundly different FA patterns are observed for M4 cells than for MDA-MB-231 cells. At the interface between an M4 cell and the bottom ECM, large FA complexes are formed at the front of the cell protrusions and also in the grooves between adjacent rows of sawteeth (Figure 6E; paxillin channel). These FAs display a maximum intensity at a z of approximately 1.4 μm (Figure 6F, 1–3 blue dashed lines, and Figure S3C, top panel). Above z = 2.0 μm, the M4 cells exhibit organized FA patterns surrounding each sawtooth, resulting in brighter outlines of the sawteeth (Figure 6E; paxillin channel). Furthermore, thick actin structures are seen around the sawteeth through the entire z section (Figure 6E; actin channel). This phenomenon is visible in a 3D reconstruction, which clearly shows the outlines of sawteeth (Movie S6). In contrast to FAs in MDA-MB-231 cells, FAs of M4 cells interact with the ECM on the side walls of the sawteeth (Figure 6F; 1− 3 y−z view). Furthermore, the FA complexes are located along the slopes of the sawteeth, extend from the bottoms to the tops of the sawteeth (Figure 6F, 4−6 x−z view, and Movie S6). This finding is further substantiated by the quantification of fluorescence intensity, which shows a broad distribution over the entire z depth (Figure S3C; bottom panel). The y−z view of FAs 4−6 further illustrates that FA complexes are assembled on the side walls of the sawteeth for a range of z values. SEM images provide further evidence that the cortical plasticity of M4 cells is distinct from that of MDA-MB-231 cells (Figure 6G). Multiple protrusions are formed around M4 cells, compared with only one lamellipodium for MDA-MB-231 cells (Figure 6C,G). The cell boundary of M4 cells is also highly dynamic, with extensive membrane ruffling and blebbing (Figure 6G). Zooming in on one dominant protrusion indicates that the cell boundary, especially at the front of the protrusion, is able to wrap around individual sawteeth (Figure 6G; see white arrows in the zoomed-in image). This observation is consistent with the ability of M4 cells to assemble FA complexes surrounding sawteeth. Based on these experimental results, we summarize the primary differences in actin cytoskeleton and FA patterns between MDA-MB-231 and M4 cells on sawteeth in a schematic (Figure 6D,H). For MDA-MB-231 cells, the high cortical rigidity limits the contact of membranes to the sawtooth structures. As a result, the majority of the FAs are located on the top of the sawteeth (Figure 6D). In contrast, the distinct cortical plasticity of M4 cells allows maximum contact with the sawteeth structures and FA formation throughout the sawteeth (Figure 6H).
Although it has been shown that the application of cytoskeletal force generated by cells on the ECM directly affects FA assembly,69,70 the overall cell shape or distortion of the cell shape is also known to modulate the size and distribution of FAs.71 Here, we found that various cell types exhibit both distinct cell boundary deformability and FA organization. It has been suggested that FA size and distribution determine the adhesion strength72 and FA composition,73 spatially control the activation of downstream messengers,74 and determine the balance among ECM-activated signaling pathways.51 Thus, the distinct FA organization patterns we observed in breast cancer cells migrating on sawteeth may play an important regulatory role. Topography-induced membrane curvature could represent another critical factor. Indeed, instead of being passively determined by the actin cytoskeleton, membrane shape, and deformation has been shown to actively regulate the recruitment of curvature-sensitive proteins and activators of actin polymerization.75,76 The recruited curvature-sensitive proteins can further promote membrane deformation; however, the triggered actin polymerization limits this positive feedback process by stiffening the cortex.75,76 This antagonistic interplay generates propagating waves of both curvature sensing proteins and actin polymerization with changes in local membrane curvature.75–77 In addition, it has been determined that out-of-plane curvature provided by microscale wires or tubules also regulates the collective behavior of epithelial cell monolayer.78,79 This raises the possibility that the different membrane deformability we observed in MDA-MB-231 and M4 cells results in the distinct recruitment of curvature-sensing proteins and actin nucleators, thereby leading to the generation of actin polymerization waves in opposite directions. We hypothesize that differences in the balance between cortex deformation and FA formation ability in MDA-MB-231 and M4 cells promote distinct cytoskeletal dynamics and migration behaviors.
CONCLUSIONS
Contact guidance plays a critical role in cancer invasion and metastatic processes. Using multiple breast cancer cell lines and symmetric and asymmetric nanotopographic designs, we examined the biophysical and molecular mechanisms behind contact guidance and directional bias. We showed that all of the breast cancer cell lines are able to sense and respond to physical guidance cues, yet they do so in distinct ways. We envision that this behavior is dependent on distinct intrinsic characteristics of the cancer cell lines. Consistent with prior findings, the bias in cell migration we measured is in the same direction as, and likely arises from, asymmetries in the direction of actin polymerization. Furthermore, we identified distinct local cell cortical plasticity and heterogeneity in FA patterns as key differences between breast cancer cell types. We hypothesize that this difference in FA patterns and the associated difference in the deformation ability of cell boundaries into the valleys of the textures are both driven by, and ultimately regulate, the bias in actin polymerization and cell migration. Because we can average observations over identical nanotopographic elements (Figure 5), our approach is well suited for in-depth investigations of intracellular signaling to identify the molecular mechanisms underlying the distinct FA patterns, actin dynamics, and cell boundary shapes.
MATERIALS AND METHODS
Surface Fabrication and Replication.
The nanoridge and sawtooth topographies that were employed to study migratory and cytoskeletal responses were designed and fabricated using multiphoton absorption polymerization (MAP). More details regarding this method can be found elsewhere.80,81 Briefly, the output of an ultrafast Ti:sapphire laser (Coherent Mira 900) tuned to 800 nm was passed through a high-numerical-aperture objective (Zeiss alpha-Plan Fluar 100×; numerical aperture 1.45) and focused onto a photopolymerizable resin ((1:1 w/w tris (2-hydroxy ethyl) isocyanurate triacrylate (SR368)/ethoxylated (6) trimethylolpropane triacrylate (SR499) (both from Sartomer), 3 wt % Lucirin TPO-L (BASF)) that was sandwiched between a glass coverslip and a microscope slide. Based on the experimental conditions, the photoinitiator undergoes efficient two-photon absorption only in the focal region of the objective, allowing for the fabrication of three-dimensional structures with arbitrary shape. The sample was mounted on a motorized stage that can be controlled by a computer. A LabVIEW program (National Instruments) was used to control the stage movement to fabricate the desired the patterns. The same program controlled a shutter to dictate when and where polymerization should occur. After fabrication was completed, the sample was soaked in two containers of ethanol for 3 min each to remove the unreacted monomer resin. The MAP-fabricated “master” structure was then baked/dried in oven at 110 °C for at least 1 h.
To reproduce nanotopographic surfaces, we used solvent-assisted nanotransfer molding42 to make a negative-relief mold of the “master” structure fabricated with MAP. A film of hard PDMS42,82 was spincoated onto the original pattern. This PDMS mixture included hexanes to decrease viscosity to allow the film to conform optimally onto the structures being coated, thereby improving the resolution of the mold. After sitting at room temperature for 2 h, the patterned microscope slide and coated film were baked at 60 °C for 1 h. Sylgard 184 (10:1 w/w elastomer base/curing agent; Dow Corning) was poured on top of the baked sample to form a layer approximately 1 cm thick. The same was then returned to the oven for 70 min. Once baking was completed, the mold was peeled from the “master” in the direction parallel to the ridges. For the sawteeth mold, peeling should occur from the tail to the head of the sawteeth to prevent damage to the pattern. Molds can be used to make replicas of the original MAP structure. A drop of the same resin used for MAP was sandwiched between a functionalized coverslip and the mold, and the sandwich was exposed to UV light for 5 min. Using this process, many replicas of an original pattern can be produced in a short amount of time.
Cell Culture.
Benign epithelial cell line MCF10A (M1) and its metastatic mutant cell line MCF10CA1 (M4) from the MCF10A cell series were used (Barbara Ann Karmanos Cancer Institute, Detroit, MI). Cells were cultured in DMEM/F12 (Invitrogen, Carlsbad, CA) supplemented with 5% horse serum (Invitrogen) at 5% CO2 in humidified culture incubators, as previously described. The medium for M1 cells was additionally supplemented with 10 μg/mL insulin (Invitrogen), 10 ng/mL EGF (Peprotech, Rocky Hill, NJ), 0.5 μg/mL hydrocortisone, and 100 ng/mL cholera toxin (both from Sigma, St. Louis, MO). HS578T and BT549 cells, kind gifts from Dr. Stanley Lipkowitz, were cultured in RPMI supplemented with 10% fetal bovine serum (both from Invitrogen). MDA-MB-231 cells were cultured in DMEM/high glucose supplemented with 10% fetal bovine serum.
The pTK92_Lifeact-GFP plasmid was a gift from Iain Cheeseman (Addgene plasmid #46356). To generate M1 and MDA-MB-231 cells stably expressing Lifeact-eGFP, phoenix cells (human kidney epithelial) were transfected with the plasmid with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocol, then the cell culture medium containing retroviral particles was collected at 48 h after transfection. After the collected medium was filtered with 0.22 μm filters, viral particles were added to M1 and MDA-MB-231 cells. Stably infected cells were selected and maintained in puromycin-containing media (2.5 μg/mL).
Individual Cell Migration and Time-Lapse Imaging.
Prior to cell seeding, nanoridges or nanosawteeth surfaces were coated with 20 μg/mL collagen IV (BD Biosciences) at 4 °C for about 1 h and then mounted back to a homemade 6-well plate. Cells were trypsinized and resuspended to 1 × 104 cell/mL in each own growth medium, and then 2 mL of cell suspension was added to each well. After the cells were allowed to adhere for approximately 1 h, nonadherent cells were washed off and each well was refilled with 2 mL of fresh medium. The cells were then cultured overnight. Two hours before imaging, fresh growth medium was replaced for each cell line. For time-lapse imaging, the plate was placed in the incubator chamber (37 °C, 5% CO2) of a Zeiss Observer 2.1 microscope with an automated stage. Phase-contrast images were taken every 3 min for approximately 20 h.
Live-Cell Actin Fluorescent Imaging. Similar to individual cell migration experiments, a nanosawtooth surface was coated with 20 μg/mL collagen IV at 4 °C for about 1 h, then mounted in a homemade 35 mm Petri dish. Lifeact-eGFP/MDA-MB-231 and Lifeact-eGFP/M4 cells were harvested following the same protocol as in the individual cell migration experiments. On the second day, cells were imaged with a Zeiss LSM 880 laser-scanning confocal microscope. Both fluorescent and bright-field images were taken every 10 s using an oil-immersion, 63× objective and a zoom-in factor of 2.5.
Immunostaining.
Cells were fixed in 4% formaldehyde without (paxillin) or with (tubulin) 2 mM EGTA (bioWORLD, Dublin, OH) and 2 mM MgCl2 (Quality Biological Inc., Gaithersburg, MD) for 15 min, followed by permeabilization in 0.1% Triton X-100 for 15 min and blocking of unspecific antibody binding with 1% bovine serum albumin (BSA) for 1 h. Targets were labeled with either antitubulin (1:200, MAB1864, Millipore) or antipaxillin (1:200, 612405, BD) primary antibody in 0.1% BSA at 4 °C overnight. On the second day, the primary antibodies were detected by secondary, fluorescently labeled antibodies (1:200, Invitrogen). Meanwhile, F-actin was labeled with phalloidin-TRITC (1:200, Invitrogen), and nuclei were labeled with DAPI (Invitrogen). Super-resolution images of the specimens were taken with a Zeiss LSM 880 confocal microscope in Airyscan mode, and 3D reconstruction movies were generated using ZEN software (Zeiss, Germany). Further image analysis, such as generating x−z/y−z view images, was performed in ImageJ (NIH, Bethesda, MD).
Quantitative Analysis of Cell Migration.
After time-lapse images were postprocessed, cell tracking was performed using Manual Tracking in ImageJ, and cells were outlined manually in MATLAB (Mathworks). The files containing all cell tracks were imported into MATLAB for further analysis. Cell tracks were smoothed with a 10frame (30 min) unweighted sliding window. This period is sufficient to allow cells to move about one nuclear diameter. Using the smoothed center positions, cell velocities were determined by calculating the displacement between two frames: , where Δt is the interval between two frames. Assuming the uncertainty in finding cell center is 1 pixel (0.625 μm) for both the x and y directions, a velocity was only counted if the net displacement between two frames was above 1.4 pixels. The direction of the motion at each frame was determined by the angle between and the x axis, based on which histograms of direction of motion were created. All cell instantaneous velocities within a certain angle range relative to ridges were averaged to plot the velocity profile with respect to orientation. To calculate the mean velocity components relative to the nanosawtooth orientation for each cell, the velocity components along the x axis were separated based on their signs, with positive being the same as the nanosawtooth orientation and negative being the opposite orientation.
The velocity direction distribution was also plotted for each speed quartile and each long-term persistence tertile. The angles were determined relative to the sawtooth orientation, meaning that angles that are left−right symmetric about the sawtooth orientation are considered to be the same. This process transforms the original range of velocity direction from −180−180° to 0−180°. To estimate the variations in our distributions, we adapted a random resampling Bootstrap method.83,84 First, we randomly divided all velocities into 10 groups for each speed quartile or persistence tertile. To distribute the velocities randomly, we then selected a random seed between 20 and 232 for each of the five cell lines. The seeds are 418932850, 1196140743, 2348838240, 4112460544, and 4144164703 for MDAMB-231, HS578T, M1, BT549, and M4 cells, respectively. The velocities whose speed values are zero were discarded because their directionality cannot be determined. The velocity direction distribution was then calculated for each of the 10 groups. We considered the measurement from each of the 10 groups as independent and calculated the average velocity direction distribution and standard error of the mean from the measurements of the 10 groups.
Averaged Actin Kymograph Analysis.
We measured the mean actin fluorescence and actin dynamic around one sawtooth by averaging across all sawteeth in either a single movie or multiple movies. First, the images with both bright-field and fluorescence channels were rotated in ImageJ such that all sawteeth were aligned horizontally with their positive direction (tail to head) facing right, and the image edges were discarded. The sawtooth regions and boundaries were subtracted from the bright-field channel. Because the light aberration caused by cells resulted in the inability to recognize some sawteeth, sawteeth that appeared to be connected with others or smaller than a certain threshold were excluded from the analysis. After subtraction, the sawtooth outlines were superimposed on the actin fluorescent channel, and 51 points were placed evenly about the circumference of each outline. For each sawtooth, the sawtooth centroid was used as the origin point, and the horizontal row passing the centroid was used as the x axis with its positive direction facing right (same as the sawtooth positive direction). Then, the angle of each of the 51 points was calculated relative to the axis positive direction, and the position 0, which is the head of the sawtooth, was identified as the point with the smallest angle. The 25 points from position 0 in the clockwise direction were assigned indices 1 to 25 with position 1 near the sawtooth head and position 25 near the sawtooth tail. Similarly, the 25 points from position 0 in the counter clockwise direction were given indices −1 to −25. As the 51 points were evenly placed, positions ±1 are nearly symmetric to the sawtooth head and positions ±25 are nearly symmetric to the sawtooth tail. The actin fluorescence intensity within a constant radius around each point was averaged. Then, we aligned the 51 points linearly so that position 0 was in the middle and positions ±25 were in the two sides. To quantify the actin dynamics for two cell lines in an unbiased way, we first calculated averaged intensity for position 0 for all frames and then made a plot of intensity versus time. The time point tw was determined as the frame of the first peak, which has the average intensity above 90% of the maximum intensity in the plot. A kymograph showing the actin dynamic around one sawtooth was then generated by measuring the actin intensity profile as mentioned for 6 min before and after time point tw. The kymographs from different sawteeth were averaged by using the position 0 and time tw as the center.
Scanning Electron Microscopy Imaging.
For SEM imaging, cells were washed with 0.1 M Sorensen’s sodium phosphate buffer three times to remove any growth medium and fixed with 2.5% glutaraldehyde in the same buffer at 4 °C overnight. After being rinsed, cells were postfixed with 1% osmium tetroxide for 1 h. Then, cells were dehydrated through ethanol series of 35, 50, 85, 95, and 100% ethanol solutions for 10 min each, followed by chemically drying with hexamethyldisilazane (Sigma-Aldrich) for 5 min. The samples were then sputter-coated with gold and imaged using an AMRAY 1910 microscope.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Lipkowitz and the Parent, Losert, and Fourkas laboratory members for valuable discussions and suggestions. We also thank C.H. Stuelten and R. Lee for their help with the experimental setup and quantification of cell migration. We thank S. Garfield, L. Lim, and P. Mannan in the confocal microscopy core in CCR for their assistance with imaging. We thank J. Harrison and P. Blakely in the microscopy and image analysis laboratory at University of Michigan for the assistance with scanning electron microscopy. This work was supported by the Intramural Research Program of the Center for Cancer Research, NCI, National Institutes of Health (C.A.P.), by the NCI-UMD Partnership for Integrative Cancer Research (S.C.), by funds from the University of Michigan (S.C. and C.A.P.) and by AFOSR Grant FA9550-16-1-0052 (J.T.F. and W.L.).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b07140.
Description of the principle of optical-flow algorithm; Figures S1−S3 and Table S1, which depict additional results (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Amini-Nik S; Cambridge E; Yu W; Guo A; Whetstone H; Nadesan P; Poon R; Hinz B; Alman BA Beta-Catenin-Regulated Myeloid Cell Adhesion and Migration Determine Wound Healing. J. Clin. Invest. 2014, 124, 2599–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Friedl P; Alexander S Cancer Invasion and the Microenvironment: Plasticity and Reciprocity. Cell 2011, 147, 992–1009. [DOI] [PubMed] [Google Scholar]
- (3).Lammermann T; Afonso PV; Angermann BR; Wang JM; Kastenmuller W; Parent CA; Germain RN Neutrophil Swarms Require Ltb4 and Integrins at Sites of Cell Death in Vivo. Nature 2013, 498, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Scarpa E; Mayor R. Collective Cell Migration in Development. J. Cell Biol. 2016, 212, 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Petrie RJ; Doyle AD; Yamada KM Random Versus Directionally Persistent Cell Migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Chen S; Li N; Hsu SF; Zhang J; Lai PY; Chan CK; Chen W. Intrinsic Fluctuations of Cell Migration under Different Cellular Densities. Soft Matter 2014, 10, 3421–3425. [DOI] [PubMed] [Google Scholar]
- (7).Bagorda A; Parent CA Eukaryotic Chemotaxis at a Glance. J. Cell Sci. 2008, 121, 2621–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Parent CA Making All the Right Moves: Chemotaxis in Neutrophils and Dictyostelium. Curr. Opin. Cell Biol. 2004, 16, 4–13. [DOI] [PubMed] [Google Scholar]
- (9).Sunyer R; Conte V; Escribano J; Elosegui-Artola A; Labernadie A; Valon L; Navajas D; Garcia-Aznar JM; Munoz JJ; Roca-Cusachs P; Trepat X. Collective Cell Durotaxis Emerges from Long-Range Intercellular Force Transmission. Science 2016, 353, 1157–1161. [DOI] [PubMed] [Google Scholar]
- (10).Shanley LJ; Walczysko P; Bain M; MacEwan DJ; Zhao M. Influx of Extracellular Ca2+ Is Necessary for Electrotaxis in Dictyostelium. J. Cell Sci. 2006, 119, 4741–4748. [DOI] [PubMed] [Google Scholar]
- (11).Kim DH; Provenzano PP; Smith CL; Levchenko A. Matrix Nanotopography as a Regulator of Cell Function. J. Cell Biol. 2012, 197, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Nguyen AT; Sathe SR; Yim EK From Nano to Micro: Topographical Scale and Its Impact on Cell Adhesion, Morphology and Contact Guidance. J. Phys.: Condens. Matter 2016, 28, 183001. [DOI] [PubMed] [Google Scholar]
- (13).Doyle AD; Carvajal N; Jin A; Matsumoto K; Yamada KM Local 3d Matrix Microenvironment Regulates Cell Migration through Spatiotemporal Dynamics of Contractility-Dependent Adhesions. Nat. Commun. 2015, 6, 8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Provenzano PP; Eliceiri KW; Campbell JM; Inman DR; White JG; Keely PJ Collagen Reorganization at the Tumor Stromal Interface Facilitates Local Invasion. BMC Med. 2006, 4, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Provenzano PP; Inman DR; Eliceiri KW; Trier SM; Keely PJ Contact Guidance Mediated Three-Dimensional Cell Migration Is Regulated by Rho/Rock-Dependent Matrix Reorganization. Biophys. J. 2008, 95, 5374–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Alexander S; Koehl GE; Hirschberg M; Geissler EK; Friedl P. Dynamic Imaging of Cancer Growth and Invasion: A Modified Skin-Fold Chamber Model. Histochem. Cell Biol. 2008, 130, 1147–1154. [DOI] [PubMed] [Google Scholar]
- (17).Ray A; Slama ZM; Morford RK; Madden SA; Provenzano PP Enhanced Directional Migration of Cancer Stem Cells in 3d Aligned Collagen Matrices. Biophys. J. 2017, 112, 1023–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wolf K; Te Lindert M; Krause M; Alexander S; Te Riet J; Willis AL; Hoffman RM; Figdor CG; Weiss SJ; Friedl P. Physical Limits of Cell Migration: Control by Ecm Space and Nuclear Deformation and Tuning by Proteolysis and Traction Force. J. Cell Biol. 2013, 201, 1069–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Dvir T; Timko BP; Kohane DS; Langer R. Nanotechnological Strategies for Engineering Complex Tissues. Nat. Nanotechnol. 2011, 6, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Potthoff E; Franco D; D’Alessandro V; Starck C; Falk V; Zambelli T; Vorholt JA; Poulikakos D; Ferrari A. Toward a Rational Design of Surface Textures Promoting Endothelialization. Nano Lett. 2014, 14, 1069–1079. [DOI] [PubMed] [Google Scholar]
- (21).Ramirez-San Juan GR; Oakes PW; Gardel ML Contact Guidance Requires Spatial Control of Leading-Edge Protrusion. Mol. Biol. Cell 2017, 28, 1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ray A; Lee O; Win Z; Edwards RM; Alford PW; Kim DH; Provenzano PP Anisotropic Forces from Spatially Constrained Focal Adhesions Mediate Contact Guidance Directed Cell Migration. Nat. Commun. 2017, 8, 14923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Onesto V; Cancedda L; Coluccio ML; Nanni M; Pesce M; Malara N; Cesarelli M; Di Fabrizio E; Amato F; Gentile F. Nano-Topography Enhances Communication in Neural Cells Networks. Sci. Rep. 2017, 7, 9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Teo BK; Wong ST; Lim CK; Kung TY; Yap CH; Ramagopal Y; Romer LH; Yim EK Nanotopography Modulates Mechano transduction of Stem Cells and Induces Differentiation through Focal Adhesion Kinase. ACS Nano 2013, 7, 4785–4798. [DOI] [PubMed] [Google Scholar]
- (25).Yang K; Yu SJ; Lee JS; Lee HR; Chang GE; Seo J; Lee T; Cheong E; Im SG; Cho SW Electroconductive Nanoscale Topography for Enhanced Neuronal Differentiation and Electrophysiological Maturation of Human Neural Stem Cells. Nanoscale 2017, 9, 18737–18752. [DOI] [PubMed] [Google Scholar]
- (26).Vicente-Manzanares M; Webb DJ; Horwitz AR Cell Migration at a Glance. J. Cell Sci. 2005, 118, 4917–4919. [DOI] [PubMed] [Google Scholar]
- (27).Mitra SK; Hanson DA; Schlaepfer DD Focal Adhesion Kinase: In Command and Control of Cell Motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [DOI] [PubMed] [Google Scholar]
- (28).Kanchanawong P; Shtengel G; Pasapera AM; Ramko EB; Davidson MW; Hess HF; Waterman CM Nanoscale Architecture of Integrin-Based Cell Adhesions. Nature 2010, 468, 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Devreotes P; Horwitz AR Signaling Networks That Regulate Cell Migration. Cold Spring Harbor Perspect. Biol. 2015, 7, a005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Driscoll MK; Sun X; Guven C; Fourkas JT; Losert W. Cellular Contact Guidance through Dynamic Sensing of Nanotopography. ACS Nano 2014, 8, 3546–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Sun X; Driscoll MK; Guven C; Das S; Parent CA; Fourkas JT; Losert W. Asymmetric Nanotopography Biases Cytoskeletal Dynamics and Promotes Unidirectional Cell Guidance. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 12557–12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kim DH; Han K; Gupta K; Kwon KW; Suh KY; Levchenko A. Mechanosensitivity of Fibroblast Cell Shape and Movement to Anisotropic Substratum Topography Gradients. Biomaterials 2009, 30, 5433–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Park J; Kim HN; Kim DH; Levchenko A; Suh KY Quantitative Analysis of the Combined Effect of Substrate Rigidity and Topographic Guidance on Cell Morphology. IEEE Trans Nanobioscience 2012, 11, 28–36. [DOI] [PubMed] [Google Scholar]
- (34).Ventre M; Natale CF; Rianna C; Netti PA Topographic Cell Instructive Patterns to Control Cell Adhesion, Polarization and Migration. J. R. Soc., Interface 2014, 11, 20140687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kadota M; Yang HH; Gomez B; Sato M; Clifford RJ; Meerzaman D; Dunn BK; Wakefield LM; Lee MP Delineating Genetic Alterations for Tumor Progression in the Mcf10a Series of Breast Cancer Cell Lines. PLoS One 2010, 5, No. e9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Marella NV; Malyavantham KS; Wang J; Matsui S; Liang P; Berezney R. Cytogenetic and Cdna Microarray Expression Analysis of Mcf10 Human Breast Cancer Progression Cell Lines. Cancer Res. 2009, 69, 5946–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Tang B; Vu M; Booker T; Santner SJ; Miller FR; Anver MR; Wakefield LM Tgf-Beta Switches from Tumor Suppressor to Prometastatic Factor in a Model of Breast Cancer Progression. J. Clin. Invest. 2003, 112, 1116–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Foulkes WD; Smith IE; Reis-Filho JS Triple-Negative Breast Cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [DOI] [PubMed] [Google Scholar]
- (39).Santner SJ; Dawson PJ; Tait L; Soule HD; Eliason J; Mohamed AN; Wolman SR; Heppner GH; Miller FR Malignant Mcf10ca1 Cell Lines Derived from Premalignant Human Breast Epithelial Mcf10at Cells. Breast Cancer Res. Treat. 2001, 65, 101–110. [DOI] [PubMed] [Google Scholar]
- (40).Song KH; Kwon KW; Choi JC; Jung J; Park Y; Suh KY; Doh J. T Cells Sense Biophysical Cues Using Lamellipodia and Filopodia to Optimize Intraluminal Path Finding. Integr Biol. (Camb) 2014, 6, 450–459. [DOI] [PubMed] [Google Scholar]
- (41).Tan J; Saltzman WM Topographical Control of Human Neutrophil Motility on Micropatterned Materials with Various Surface Chemistry. Biomaterials 2002, 23, 3215–3225. [DOI] [PubMed] [Google Scholar]
- (42).Sun X; Hourwitz MJ; Baker EM; Schmidt BUS; Losert W; Fourkas JT Replication of Biocompatible, Nanotopographic Surfaces. Sci. Rep. 2018, 8, 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Arocena M; Rajnicek AM; Collinson JM Requirement of Pax6 for the Integration of Guidance Cues in Cell Migration. R. Soc. Open Sci. 2017, 4, 170625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Tzvetkova-Chevolleau T; Stephanou A; Fuard D; Ohayon J; Schiavone P; Tracqui P. The Motility of Normal and Cancer Cells in Response to the Combined Influence of the Substrate Rigidity and Anisotropic Microstructure. Biomaterials 2008, 29, 1541–1551. [DOI] [PubMed] [Google Scholar]
- (45).Friedl P. Prespecification and Plasticity: Shifting Mechanisms of Cell Migration. Curr. Opin. Cell Biol. 2004, 16, 14–23. [DOI] [PubMed] [Google Scholar]
- (46).Friedl P; Wolf K. Plasticity of Cell Migration: A Multiscale Tuning Model. J. Cell Biol. 2010, 188, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Kao J; Salari K; Bocanegra M; Choi YL; Girard L; Gandhi J; Kwei KA; Hernandez-Boussard T; Wang P; Gazdar AF; Minna JD; Pollack JR Molecular Profiling of Breast Cancer Cell Lines Defines Relevant Tumor Models and Provides a Resource for Cancer Gene Discovery. PLoS One 2009, 4, No. e6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bear JE; Haugh JM Directed Migration of Mesenchymal Cells: Where Signaling and the Cytoskeleton Meet. Curr. Opin. Cell Biol. 2014, 30, 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Park J; Kim DH; Levchenko A. Topotaxis: A New Mechanism of Directed Cell Migration in Topographic Ecm Gradients. Biophys. J. 2018, 114, 1257–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Kim DH; Seo CH; Han K; Kwon KW; Levchenko A; Suh KY Guided Cell Migration on Microtextured Substrates with Variable Local Density and Anisotropy. Adv. Funct. Mater. 2009, 19, 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Park J; Kim DH; Kim HN; Wang CJ; Kwak MK; Hur E; Suh KY; An SS; Levchenko A. Directed Migration of Cancer Cells Guided by the Graded Texture of the Underlying Matrix. Nat. Mater. 2016, 15, 792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Comelles J; Caballero D; Voituriez R; Hortigüela, V.; Wollrab, V.; Godeau, A. L.; Samitier, J.; Martinez, E.; Riveline, D. Cells as Active Particles in Asymmetric Potentials: Motility under External Gradients. Biophys. J. 2014, 107, 1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Caballero D; Voituriez R; Riveline D. Protrusion Fluctuations Direct Cell Motion. Biophys. J. 2014, 107, 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Caballero D; Voituriez R; Riveline D. The Cell Ratchet: Interplay between Efficient Protrusions and Adhesion Determines Cell Motion. Cell Adh Migr 2015, 9, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Caballero D; Comelles J; Piel M; Voituriez R; Riveline D. Ratchetaxis: Long-Range Directed Cell Migration by Local Cues. Trends Cell Biol. 2015, 25, 815–827. [DOI] [PubMed] [Google Scholar]
- (56).Maiuri P; Rupprecht JF; Wieser S; Ruprecht V; Benichou O; Carpi N; Coppey M; De Beco S; Gov N; Heisenberg CP; Lage Crespo C; Lautenschlaeger F; Le Berre M; Lennon-Dumenil AM; Raab M; Thiam HR; Piel M; Sixt M; Voituriez R. Actin Flows Mediate a Universal Coupling between Cell Speed and Cell Persistence. Cell 2015, 161, 374–386. [DOI] [PubMed] [Google Scholar]
- (57).Bettinger CJ; Orrick B; Misra A; Langer R; Borenstein JT Microfabrication of Poly (Glycerol-Sebacate) for Contact Guidance Applications. Biomaterials 2006, 27, 2558–2565. [DOI] [PubMed] [Google Scholar]
- (58).Oakley C; Brunette DM The Sequence of Alignment of Microtubules, Focal Contacts and Actin Filaments in Fibroblasts Spreading on Smooth and Grooved Titanium Substrata. J. Cell Sci. 1993, 106, 343–354. [DOI] [PubMed] [Google Scholar]
- (59).Ezratty EJ; Bertaux C; Marcantonio EE; Gundersen GG Clathrin Mediates Integrin Endocytosis for Focal Adhesion Disassembly in Migrating Cells. J. Cell Biol. 2009, 187, 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Ezratty EJ; Partridge MA; Gundersen GG Microtubule Induced Focal Adhesion Disassembly Is Mediated by Dynamin and Focal Adhesion Kinase. Nat. Cell Biol. 2005, 7, 581–590. [DOI] [PubMed] [Google Scholar]
- (61).Weiner OD; Marganski WA; Wu LF; Altschuler SJ; Kirschner MW An Actin-Based Wave Generator Organizes Cell Motility. PLoS Biol. 2007, 5, No. e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Doubrovinski K; Kruse K. Cell Motility Resulting from Spontaneous Polymerization Waves. Phys. Rev. Lett. 2011, 107, 258103. [DOI] [PubMed] [Google Scholar]
- (63).Kruse K. Cell Crawling Driven by Spontaneous Actin Polymerization Waves In Physical Models of Cell Motility; Aranson IS, Ed.; Springer: Cham, 2016; pp 69–93. [Google Scholar]
- (64).Bretschneider T; Diez S; Anderson K; Heuser J; Clarke M; Muller-Taubenberger A; Kohler J; Gerisch G. Dynamic Actin Patterns and Arp2/3 Assembly at the Substrate-Attached Surface of Motile Cells. Curr. Biol. 2004, 14, 1–10. [DOI] [PubMed] [Google Scholar]
- (65).Ohara PT; Buck RC Contact Guidance in Vitro. A Light, Transmission, and Scanning Electron Microscopic Study. Exp. Cell Res. 1979, 121, 235–249. [DOI] [PubMed] [Google Scholar]
- (66).Kubow KE; Shuklis VD; Sales DJ; Horwitz AR Contact Guidance Persists under Myosin Inhibition Due to the Local Alignment of Adhesions and Individual Protrusions. Sci. Rep. 2017, 7, 14380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Zimerman B; Arnold M; Ulmer J; Blummel J; Besser A; Spatz JP; Geiger B. Formation of Focal Adhesion-Stress Fibre Complexes Coordinated by Adhesive and Non-Adhesive Surface Domains. IEE Proc.: Nanobiotechnol. 2004, 151, 62–66. [DOI] [PubMed] [Google Scholar]
- (68).Deakin NO; Turner CE Paxillin Comes of Age. J. Cell Sci. 2008, 121, 2435–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Balaban NQ; Schwarz US; Riveline D; Goichberg P; Tzur G; Sabanay I; Mahalu D; Safran S; Bershadsky A; Addadi L; Geiger B. Force and Focal Adhesion Assembly: A Close Relationship Studied Using Elastic Micropatterned Substrates. Nat. Cell Biol. 2001, 3, 466–472. [DOI] [PubMed] [Google Scholar]
- (70).Galbraith CG; Yamada KM; Sheetz MP The Relationship between Force and Focal Complex Development. J. Cell Biol. 2002, 159, 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Chen CS; Alonso JL; Ostuni E; Whitesides GM; Ingber DE Cell Shape Provides Global Control of Focal Adhesion Assembly. Biochem. Biophys. Res. Commun. 2003, 307, 355–361. [DOI] [PubMed] [Google Scholar]
- (72).Gallant ND; Michael KE; Garcia AJ Cell Adhesion Strengthening: Contributions of Adhesive Area, Integrin Binding, and Focal Adhesion Assembly. Mol. Biol. Cell 2005, 16, 4329–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Goffin JM; Pittet P; Csucs G; Lussi JW; Meister JJ; Hinz B. Focal Adhesion Size Controls Tension-Dependent Recruitment of Alpha-Smooth Muscle Actin to Stress Fibers. J. Cell Biol. 2006, 172, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Xia N; Thodeti CK; Hunt TP; Xu Q; Ho M; Whitesides GM; Westervelt R; Ingber DE Directional Control of Cell Motility through Focal Adhesion Positioning and Spatial Control of Rac Activation. FASEB J. 2008, 22, 1649–1659. [DOI] [PubMed] [Google Scholar]
- (75).Wu Z; Su M; Tong C; Wu M; Liu J. Membrane Shape-Mediated Wave Propagation of Cortical Protein Dynamics. Nat. Commun. 2018, 9, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Yang Y; Wu M. Rhythmicity and Waves in the Cortex of Single Cells. Philos. Trans. R. Soc., B 2018, 373, 20170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Gov NS Guided by Curvature: Shaping Cells by Coupling Curved Membrane Proteins and Cytoskeletal Forces. Philos. Trans. R. Soc., B 2018, 373, 20170115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Xi W; Sonam S; Lim CT; Ladoux B. Tubular Microscaffolds for Studying Collective Cell Migration. Methods Cell Biol. 2018, 146, 3–21. [DOI] [PubMed] [Google Scholar]
- (79).Yevick HG; Duclos G; Bonnet I; Silberzan P. Architecture and Migration of an Epithelium on a Cylindrical Wire. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 5944–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).LaFratta CN; Fourkas JT; Baldacchini T; Farrer RA Multiphoton Fabrication. Angew. Chem., Int. Ed. 2007, 46, 6238–6258. [DOI] [PubMed] [Google Scholar]
- (81).Baldacchini T. Three-Dimensional Microfabrication Using Two- Photon Polymerization; William Andrew: Oxford, UK, 2016. [Google Scholar]
- (82).Kang H; Lee J; Park J; Lee HH An Improved Method of Preparing Composite Poly (Dimethylsiloxane) Moulds. Nanotechnology 2006, 17, 197. [Google Scholar]
- (83).Efron B. Bootstrap Methods: Another Look at the Jack-Knife. Ann. Stat 1979, 7, 1–26. [Google Scholar]
- (84).Efron B; Tibshirani RJ An Introduction to the Bootstrap; Chapman & Hall: New York, 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.