Abstract
Background
Obsessive-compulsive disorder (OCD) is a debilitating neuropsychiatric disorder with a genetic risk component, yet identification of high-confidence risk genes has been challenging. In recent years, risk gene discovery in other complex psychiatric disorders has been achieved by studying rare de novo (DN) coding variants.
Methods
We performed whole-exome sequencing in 222 OCD parent-child trios (184 trios after quality control), comparing DN variant frequencies to 777 previously sequenced unaffected trios. We estimated the contribution of DN mutations to OCD risk and the number of genes involved. Finally, we looked for gene enrichment in other datasets and canonical pathways.
Results
DN likely gene disrupting and predicted damaging missense variants are enriched in OCD probands (RR 1.52, p=0.0005) and contribute to risk. We identified two high-confidence risk genes, each containing two DN damaging variants in unrelated probands: CHD8 and SCUBE1. We estimate that 34% of DN damaging variants in OCD contribute to risk, and that DN damaging variants in approximately 335 genes contribute to risk in 22% of OCD cases. Furthermore, genes harboring DN damaging variants in OCD are enriched for those reported in neurodevelopmental disorders, particularly Tourette’s disorder and autism spectrum disorders. An exploratory network analysis reveals significant functional connectivity and enrichment in canonical pathways, biological processes, and disease networks.
Conclusions
Our findings show a pathway toward systematic gene discovery in OCD via identification of damaging DN variants. Sequencing larger cohorts of OCD parent-child trios will reveal more OCD risk genes and provide needed insights into underlying disease biology.
Keywords: obsessive-compulsive disorder, whole-exome sequencing, CHD8, SCUBE1, autism, Tourette
INTRODUCTION
Obsessive-compulsive disorder (OCD) is an often-disabling neuropsychiatrie disorder with onset typically during adolescence or young adulthood and a lifetime prevalence of 1.5–2.5% (1–5). Obsessions are intrusive thoughts, images, or urges experienced as irrational, excessive, and accompanied by anxiety or discomfort. Compulsions are behaviors undertaken to mitigate obsessions or subjective feelings (i.e., the need to relieve a tactile sensation or achieve a “just right” feeling); they are usually repetitive, stereotyped, and excessive (6, 7). The anxiety or distress associated with obsessions and compulsions and the time spent on them are sources of lifelong morbidity in OCD, having profound negative effects on both patients’ and families’ quality of life. Symptoms can be so disabling that the World Health Organization has ranked OCD among the 10 most debilitating disorders of any kind, in terms of lost earnings and diminished quality of life (8, 9). Furthermore, OCD has been linked to significantly increased mortality, even after controlling for comorbid psychiatric conditions, which can occur in up to 75% of cases (10, 11). Treatment-refractory disease is common, with about 40% of patients resistant to current pharmacological and psychotherapeutic treatments, and untreated OCD generally persists and becomes chronic (12, 13). The causes and underlying biology of OCD are not well understood, which has limited the development of new treatments and interventions. For these reasons, there is an urgent need for more research to elucidate OCD risk factors and disease mechanisms.
Twin and family studies provide strong evidence for a substantial genetic contribution to OCD risk, with modern estimates of heritability around 40–50% (14–17), yet progress in identifying risk genes has been slow. Decades of linkage, common-variant candidate gene association studies, and more recent genome-wide association studies in OCD (18–20) have yielded few reproducible associations and therefore have provided limited insights into disease biology. Further efforts are clearly needed to identify specific OCD risk variants and to confirm vulnerability pathways by modern genome-wide and comprehensive variant discovery approaches.
In contrast, genetic research into several other neuropsychiatric disorders has seen significant advancement in recent years. This progress is partly attributable to increasing attention toward the contribution of rare genetic sequence variation, especially de novo variants which arise spontaneously in parental germ cells or in a zygote shortly after conception. This approach has shown great success for systematic risk gene discovery in other genetically complex neuropsychiatric disorders (21–25), particularly autism spectrum disorders (26–29). While an individual rare variant is unlikely to explain a sizeable fraction of disease risk in the context of a heterogeneous genetic architecture, concurrent investigations of multiple genes implicated by rare sequence and structural variation highlight convergence toward a limited number of important underlying biological mechanisms (30). Therefore, there is a proven and reliable approach toward risk gene discovery in complex neuropsychiatric disorders that has yet to be fully leveraged in OCD.
Following these previous studies in other disorders and our pilot study suggesting a role for de novo single nucleotide variants (SNVs) in OCD risk (31), we performed whole-exome sequencing (WES) in 222 OCD parent-child trios to identify de novo SNVs and insertion-deletion variants (indels). In 184 OCD trios passing quality control, we find strong evidence for the contribution of de novo likely gene disrupting (LGD; creation or loss of a stop codon, canonical splice site, or a frameshift indel) as well as predicted damaging missense (Mis-D) variants to OCD. Furthermore, we identify two high-confidence candidate risk genes based on observing gene-level recurrence of de novo damaging (LGD + Mis-D) variants in unrelated probands: CHD8 (Chromodomain Helicase DNA Binding Protein 8) and SCUBE1 (Signal Peptide, CUB Domain And EGF Like Domain Containing 1). We estimate that 22% of OCD cases will harbor a de novo damaging SNV or indel mediating OCD risk, and that there are approximately 335 genes affected by such variants contributing to the risk. Finally, we detect significant overlap between genes with damaging de novo variants in OCD and those previously reported in Tourette’s disorder and autism.
METHODS AND MATERIALS
Subjects
This study was approved by the local institutional review boards of all participating institutions, and appropriate informed consent was obtained from participating subjects. 222 parent-child trios (139 male, 83 female), consisting of offspring meeting criteria for the diagnosis of obsessive-compulsive disorder, as defined by the Diagnostic and Statistical Manual for Mental Disorders (DSM-IV-TR or DSM-5) (32, 33), and their unaffected parents, were recruited for DNA sequencing. Trios were recruited at three sites: the University of Sao Paulo School of Medicine Obsessive-Compulsive Spectrum Disorders Program (42 trios), Centre for Addiction and Mental Health and the Frederick W. Thompson Anxiety Disorders Centre at the Sunnybrook Health Sciences Centre in Toronto (77 trios), and Yale University School of Medicine (61 trios). Additionally, we included 42 trios with OCD and chronic tics that were recruited for a separate study by TIC Genetics (25, 34). All subjects were assessed using the Structured Clinical Interview for DSM Axis I Disorders (SCID-I) (35). Subjects with a diagnosis of schizophrenia, schizoaffective disorder, autistic disorder, pervasive developmental disorder not otherwise specified, or intellectual disability were excluded from the present study. Other diagnostic criteria included: onset of symptoms prior to age 18 years; no previously diagnosed neurological disorder or OCD occurring exclusively in the context of depression; no known history of OCD in first degree relatives. Final diagnostic status was assigned based on the consensus of an experienced interviewer and a psychiatrist or psychologist after independent review and administration of the SCID. We prioritized the study of simplex OCD trios to increase the likelihood of detecting de novo sequence and structural variants. Available phenotype information, including gender and parental age, is included in Table S1.
Whole-exome sequencing (WES)
Exome capture and sequencing of blood-derived DNA from 222 affected children and their parents (666 samples total) were performed at the Yale Center for Genomic Analysis (YCGA), using the NimbleGen SeqCap EZExomeV2 (109 trios) or MedExome (113 trios) capture libraries (Roche NimbleGen, Madison, WI, USA) and the Illumina HiSeq 2000 platform (74 bp paired-end reads; Illumina, San Diego, CA). We multiplexed six samples during each capture reaction and sequencing lane, pooling parents and probands when possible. WES data from 855 unaffected parent-child trios (2565 samples total) were obtained from the Simons Simplex Collection (SSC) via the NIH Data Archive (https://ndar.nih.gov/edit_collection.html?id=2042). These control trios are comprised of unaffected siblings of autism probands from the SSC and their parents; these siblings and their parents have no evidence of autism spectrum or other neurodevelopmental disorders (36). Like our OCD samples, control WES was from blood-derived DNA and sequenced on the Illumina HiSeq 2000 sequencing platform after capture with the NimbleGen SeqCap EZExomeV2 library.
Sequence alignment, quality control, and variant calling
Alignment and variant calling of the sequencing reads followed the latest Genome Analysis Toolkit (GATK) (37) Best Practices guidelines. Details are provided in Supplemental Methods.
Mutation rate analysis
Within each cohort, we calculated the rates of de novo and inherited mutations per base pair. For accurate rate calculation, we first determined the number of “callable” base pairs per family using the GATK DepthOfCoverage tool. See Supplemental Methods for details. We compared de novo mutation rates in cases versus controls (burden analysis) using a one-tailed rate ratio test in R (https://cran.r-project.org/package=rateratio.test), considering only those variants present with a frequency of <0.01 in the ExAC v0.3.1 database (38). We compared inherited mutation rates in a similar manner but considered only those variants seen once across all cases and controls, and not reported in ExAC. See Supplemental Methods.
TADA analysis
Prior exome analyses demonstrated that the observation of even a small number of rare de novo mutations in the same gene among unrelated individuals can provide considerable statistical power to establish association (39). We used the Transmitted And De novo Association (TADA) test as a statistical method for risk gene discovery based on gene-level recurrence of de novo and inherited mutations within the classes of variants that we found enriched in OCD (29, 40). Parameter calculations and a detailed description of the method are given in Supplemental Methods.
Estimation of number of risk genes
We first used a maximum likelihood estimation (MLE) method to estimate the number of genes contributing risk to OCD, based on vulnerability to de novo damaging variants (41). See Supplemental Methods. Next, we used an alternate method for estimating the number of risk genes, using a statistical method based on the “unseen species” problem (39). See Supplemental Methods for details of these calculations.
Estimation of future risk gene discovery
Based on the predicted number of OCD risk genes, we performed simulations to predict the likely future gene discovery yield as additional OCD trios are investigated by WES. See Supplemental Methods for details of these calculations.
Gene set overlap
We used DNENRICH (42) (https://psychgen.u.hpc.mssm.edu/dnenrich/) to test whether OCD genes harboring de novo damaging mutations (89 genes; excluding two genes, TTN and CACNA1E, found to harbor de novo damaging variants in control subjects) were significantly enriched among previously reported genes identified in autism (ASD), schizophrenia (SCZ), developmental disorders (DD), Tourette’s disorder (TD), and intellectual disability (ID). Details about the gene lists and DNENRICH parameters are provided in the Supplemental Methods.
Exploratory pathway and network analyses
To determine whether all genes harboring de novo damaging variants in OCD are enriched for specific biological pathways, we used the same gene list from our gene set overlap analysis (n=89) to identify the most significant canonical pathways, biological processes, and diseases suggested by MetaCore (Clarivate Analytics) and Ingenuity Pathway Analysis (IPA, Qiagen Bioinformatics). Details of the settings used for each of these tools is given in the Supplemental Methods.
Using the GeNets algorithm (https://apps.broadinstitute.org/genets), we mapped all 89 genes harboring de novo damaging mutations in OCD onto the GeNets Metanetwork v1.0 to determine whether they are functionally connected. Details about the databases and statistical comparisons are provided in Supplemental Methods.
RESULTS
Damaging de novo SNVs and indels are associated with OCD risk
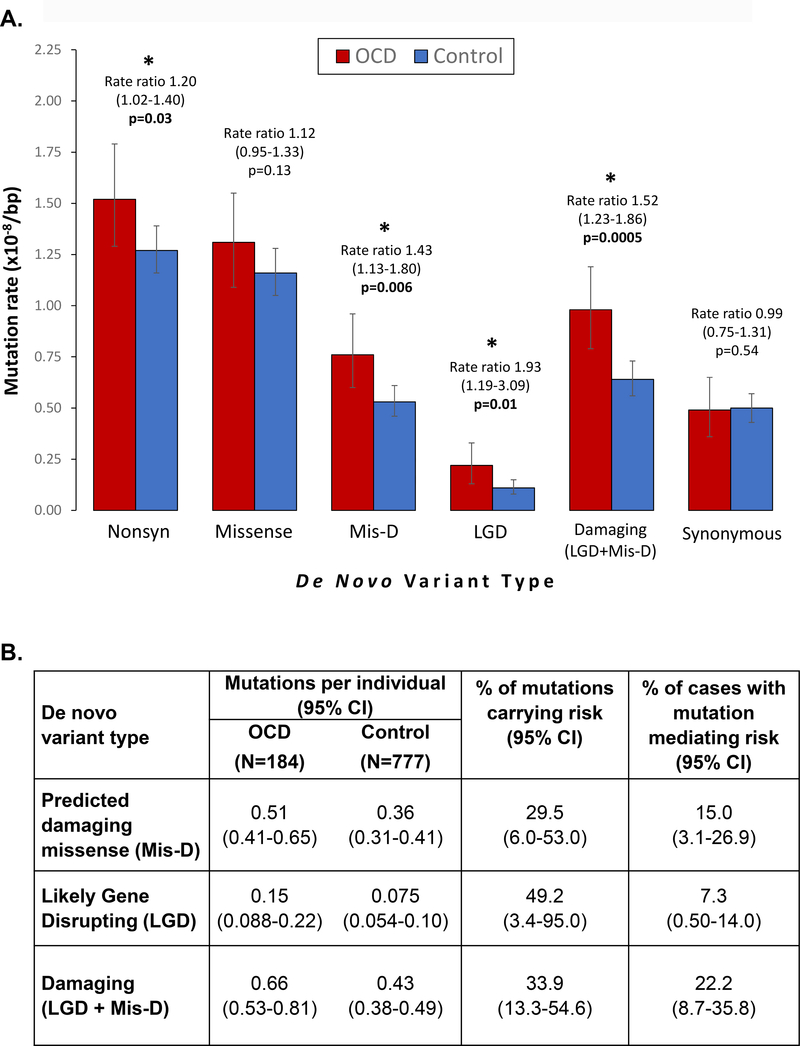
Exome sequencing was performed on 222 OCD parent-child trios. WES data from 855 unaffected trios, already sequenced from the Simons Simplex Collection, were pooled with our OCD trios for joint variant calling. After quality control methods, our sample size for a burden analysis was 184 OCD and 777 unaffected trios (Figure 1, Table 1, Table S1). To compare the de novo and inherited mutation rates between cases and controls, we limited our analysis to loci with at least 20x coverage in all members of a trio, as this was our pre-defined threshold for calling a variant (see Methods). Based on our OCD pilot study (31) and work in other neurodevelopmental disorders (22, 24, 26, 28, 43), we expected to find an enrichment of de novo LGD variants (stop codon, frameshift, or canonical splice-site variants) in OCD probands versus controls. We found a statistically significant increased rate of de novo LGD variants in OCD cases, confirming our hypothesis (rate ratio [RR] 1.93, 95% Confidence Interval [CI] 1.19–3.09, p=0.01). Furthermore, de novo missense variants predicted to be damaging by PolyPhen2 (Mis-D; Polyphen2 HDIV score >0.957) were also over-represented in OCD probands (RR 1.43, CI 1.13–1.80, p=0.006). Taken together, damaging de novo coding variants (LGD and Mis-D) occur more often in OCD probands versus controls (RR 1.52, CI 1.23–1.86, p=0.0005). We did not detect a difference in mutation rates for de novo synonymous variants (RR 0.99, CI 0.75–1.31, p=0.5) (Table 1, Figure 2A, Table S2). We did not detect a difference in mutation rates for any class of inherited variants (Tables S3–S4).
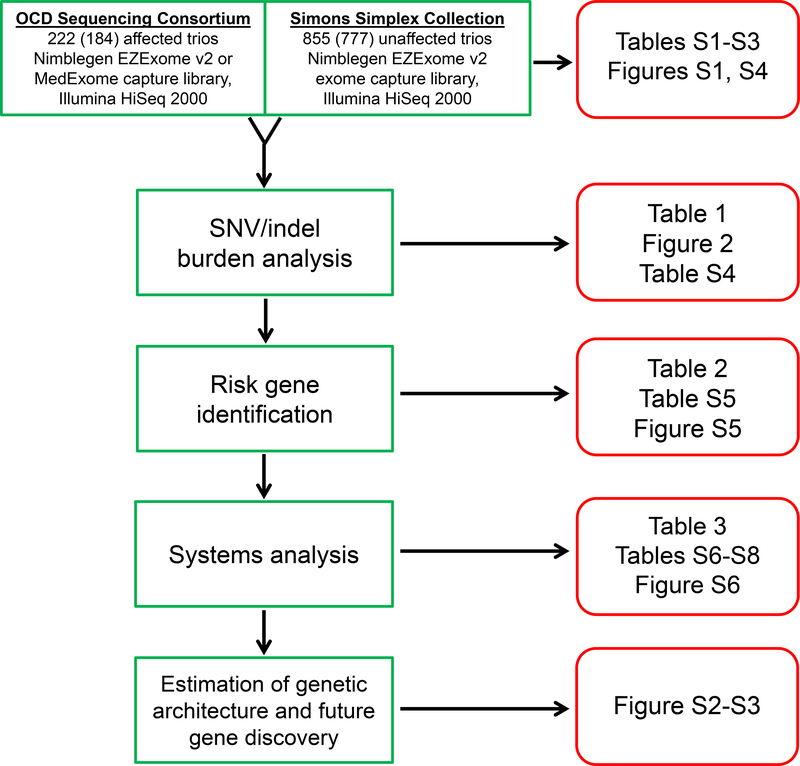
Figure 1 – Study summary.
We performed whole exome sequencing on 222 OCD and 855 control parent-child trios. After quality control, 184 OCD and 777 control trios remained for subsequent analyses. Burden analyses compared the rates of de novo and inherited single nucleotide (SNVs) and insertion-deletion (indel) variants between cases and controls. Next, we used the TADA algorithm to assess the significance of gene-level recurrence of damaging variants in our OCD group, identifying two high-confidence risk genes. Exploratory network, pathway, and cross-disorder analyses were then performed using genes harboring de novo damaging variants in our OCD subjects. Finally, based on the number of de novo damaging variants in OCD versus controls, we estimated the number of genes contributing to OCD risk, and used this estimate to predict future risk gene discovery as additional OCD parent-child trios are studied by exome sequencing.
Table 1 –
Distribution of de novo variants in OCD cases and controls
| De novo variant typea | Variant counts | Mutation rate (×10−8) per bp (95% CI)j | Estimated coding variants per individual (95% CI)k | Rate ratio (95% CI) | p-valuel | |||
|---|---|---|---|---|---|---|---|---|
| OCD (N=184) | Control (N=777) | OCD (N=184) | Control (N=777) | OCD (N=184) | Control (N=777) | |||
| Allb | 207 | 701 | 2.02 (1.75–2.31) | 1.80 (1.67–1.94) | 1.37 (1.18–1.56) | 1.22 (1.13–1.31) | 1.12 (0.95–1.31) | 0.11 |
| Codingc | 200 | 662 | 2.06 (1.78–2.36) | 1.80 (1.67–1.95) | 1.39 (1.20–1.60) | 1.22 (1.13–1.32) | 1.14 (0.99–1.30) | 0.06 |
| Synonymous SNV | 48 | 182 | 0.49 (0.36–0.65) | 0.50 (0.43–0.57) | 0.33 (0.24–0.44) | 0.34 (0.29–0.39) | 0.99 (0.75–1.31) | 0.54 |
| Nonsynonymousd | 148 | 467 | 1.52 (1.29–1.79) | 1.27 (1.16–1.39) | 1.03 (0.87–1.21) | 0.86 (0.78–0.94) | 1.20 (1.02–1.40) | 0.03 |
| All Missense (Mis) | 127 | 426 | 1.31 (1.09–1.55) | 1.16 (1.05–1.28) | 0.89 (0.74–0.20) | 0.78 (0.71–0.87) | 1.12 (0.95–1.33) | 0.13 |
| Mis-De | 74 | 195 | 0.76 (0.60–0.96) | 0.53 (0.46–0.61) | 0.51 (0.41–0.65) | 0.36 (0.31–0.41) | 1.43 (1.13–1.80) | 0.006 |
| MIs-Pf | 18 | 79 | 0.19 (0.11–0.29) | 0.22 (0.17–0.27) | 0.13 (0.074–0.20) | 0.15 (0.12–0.18) | 0.86 (0.53–1.34) | 0.76 |
| Mis-Bg | 33 | 147 | 0.34 (0.23–0.48) | 0.40 (0.34–0.47) | 0.23 (0.16–0.32) | 0.27 (0.23–0.32) | 0.85 (0.60–1.17) | 0.83 |
| Likely Gene Disrupting (LGD)h | 21 | 41 | 0.22 (0.13–0.33) | 0.11 (0.080–0.15) | 0.15 (0.088–0.22) | 0.074 (0.054–0.10) | 1.93 (1.19–3.09) | 0.01 |
| Damaging (LGD + Mis-D) | 95 | 236 | 0.98 (0.79–1.19) | 0.64 (0.56–0.73) | 0.66 (0.53–0.81) | 0.43 (0.38–0.49) | 1.52 (1.23–1.86) | 0.0005 |
| LGD SNV | 14 | 20 | 0.14 (0.079–0.24) | 0.055 (0.033–0.084) | 0.095 (0.053–0.16) | 0.037 (0.022–0.057) | 2.64 (1.39–4.93) | 0.006 |
| LGD frameshift indel | 7 | 21 | 0.072 (0.029–0.15) | 0.057 (0.035–0.088) | 0.049 (0.020–0.10) | 0.039 (0.024–0.060) | 1.28 (0.53–2.72) | 0.37 |
| Nonframeshift indel | 2 | 5 | 0.021 (0.0025–0.074) | 0.014 (0.0044–0.032) | 0.014 (0.00017–0.050) | 0.0095 (0.0030–0.022) | 1.51 (0.21–7.28) | 0.44 |
| Unknowni | 2 | 8 | 0.021 (0.0025–0.074) | 0.022 (0.0094–0.043) | 0.014 (0.00017–0.050) | 0.015 (0.0064–0.029) | 0.94 (0.14–3.88) | 0.65 |
Variants were annotated with Annovar, using RefSeq hg19 gene definitions.
“All” includes coding and non-coding variants.
“Coding” variants include synonymous, nonsynonymous, nonframeshift, and those annotated as “unknown” by Annovar.
“Nonsynonymous” variants include all missense and LGD variants.
“Mis-D” are “probably damaging” missense variants with a Polyphen2 (HDIV) score ≥0.957.
Mis-P are “possibly damaging” missense variants with a Polyphen2 (HDIV) score <0.957 and ≥0.453.
Mis-B are “benign” missense variants with a Polyphen2 (HDIV) score <0.453. Two OCD missense variants and five control missense variants had no prediction by Polyphen2, but were included in the “All Missense (Mis)” variant type.
LGD variants are those altering a stop codon, canonical splice site, and frameshift indels.
“Unknown” variants are not included in the synonymous or nonsynonymous counts.
De novo mutation rates were calculated as the number of variants divided by the number of haploid “callable” bases (see Methods).
The estimated number of de novo mutations per individual was calculated by multiplying the mutation rate by the size of the RefSeq hg19 coding exome (33,828,798 bp).
Rates were compared using a one-sided rate ratio test. Rate ratios, 95% CI, and p-values that are statistically significant (p<0.05) are underlined and in bold. See also Figure 2A.
Figure 2 – De novo damaging variants are associated with OCD risk.
(A) Bar chart comparing the rates of de novo mutation types between OCD cases (red) and controls (blue). Comparisons are between per base pair (bp) mutation rates, considering only those “callable” loci in each family and cohort that meet required sequencing depth and quality scores to support high confidence de novo variant calling. Mutation rates were compared using a one-tailed rate ratio test. Statistically significant comparisons (p<0.05) are marked with asterisks. Error bars show 95% confidence intervals. (B) For the enriched classes of de novo variants, we quantified their contribution to OCD risk in two ways. First, we estimated the percentage of observed variants carrying risk by dividing the difference in rates (estimated coding variants per individual, see Table 1 and Methods) by the rate in OCD. Second, we estimated the percentage of cases with a mutation mediating risk by subtracting the proportion of controls carrying a mutation from the proportion in OCD probands carrying a mutation.
Damaging de novo SNVs and indels contribute to OCD risk in 22% of cases
Next, we estimated the fraction of observed de novo mutations that contribute to OCD risk, based on our dataset. By dividing the de novo mutation rate difference between cases and controls by the rate in cases, we estimate that 49.2% (CI 3.4–95.0%) of de novo LGD and 29.5% (CI 6.0–53.0%) of de novo Mis-D mutations contribute to OCD risk. As a group, we estimate that 33.9% (CI 13.3–54.6%) of damaging (LGD + Mis-D) de novo mutations contribute to OCD risk (Figure 2B).
We also used our data to estimate the proportion of cases harboring a de novo mutation contributing to OCD risk. By subtracting the percentage of controls from the percentage of OCD probands with at least one de novo mutation, we estimate that 15.0% (CI 3.1–26.9%) have a de novo Mis-D mutation and 7.3% (CI 0.50–14.0%) have a de novo LGD mutation mediating OCD risk. As a group, we estimate that 22.2% (CI 8.7–35.8%) of cases have a damaging de novo mutation contributing to OCD risk (Figure 2B).
Recurrent damaging de novo variants identify two candidate risk genes
Having established that de novo damaging variants occur more frequently in OCD probands, we next asked whether these variants cluster within specific genes. We identified three genes with multiple (≥2) de novo LGD or Mis-D variants in unrelated probands. Using TADA (40) and previously established false discovery rate (FDR) thresholds, two of these genes met criteria for high-confidence risk genes (q<0.1): SCUBE1 (Signal Peptide, CUB Domain And EGF Like Domain Containing 1; q=0.091) and CHD8 (Chromodomain Helicase DNA Binding Protein 8; q=0.098). A third gene, TTN (Titin), did not meet this threshold (q=0.63) (Table 2, Table S5). For SCUBE1 and CHD8, we observed one de novo Mis-D variant and one de novo canonical splice site variant each. Each of the splice site variants decrease splicing efficiency as predicted by MaxEntScan scores (44) (88% decrease from 9.94 to 1.19 for CHD8; 100% decrease from 7.75 to −0.44 for SCUBE1) (Ingenuity Variant Analysis, Qiagen Bioinformatics).
Table 2 –
Risk gene discovery in OCD
| Gene | # de novo LGD | # de novo Mis-D | # transmitted LGD | # non-transmitted LGD | # transmitted Mis-D | # non-transmitted Mis-D | p-value | q-value (FDR) |
|---|---|---|---|---|---|---|---|---|
| SCUBE1 | 1 | 1 | 0 | 0 | 0 | 0 | 2.96 × 10−6 | 0.091 |
| CHD8 | 1 | 1 | 0 | 0 | 0 | 0 | 3.57 × 10−6 | 0.098 |
| TTN | 0 | 3 | 1 | 1 | 9 | 10 | 0.0006 | 0.63 |
We used the TADA algorithm (He et al., 2013) to estimate the likelihood of observing gene-level recurrence of de novo damaging variants in three genes in unrelated individuals. Two of these genes, SCUBE1 and CHD8, exceeded the false discovery rate (FDR) threshold for high-confidence risk genes (q<0.1). Despite observing three de novo damaging variants in TTN, this gene did not meet criteria for a high-confidence or even a probable (q<0.3) risk gene, owing to its large size and high expected de novo mutation rate. See also Figure S5, Tables S2–S3, Table S5.
Approximately 335 genes contribute to OCD risk
Based on OCD proband vulnerability to de novo damaging variants in our dataset, we used two methods to estimate the number of genes contributing to OCD risk. Using a maximum likelihood estimation (MLE) method (41), we determined the most likely number of genes to be 335 (Figure S2). This agrees with an alternate method based on the “unseen species problem” (39); the estimated number of OCD risk genes using this alternate method is 317 (95% CI 190–454).
Next, we used the estimated number of OCD risk genes (n=335) to predict the likely future gene discovery yield as additional OCD trios are investigated by WES. Based upon 10,000 simulations at each cohort size, we predict discovery of the following numbers of risk genes as we sequence more OCD parent-child trios: 24 probable risk genes, including 11 high-confidence risk genes (24 / 11 genes) at 500 trios; 77 / 40 genes at 1,000 trios; 202 / 113 genes at 2,000 trios; 323 / 189 genes at 3,000 trios (Figure S3).
Overlap with TD, ASD and CHD8 target genes
Using DNENRICH (42), we found significant overlap between genes harboring de novo damaging variants in OCD (n=89, excludes occurrences in controls) and several gene sets from the literature (Table 3, Table S6). Our OCD genes were significantly enriched for genes harboring de novo nonsynonymous (LGD, missense) variants in Tourette’s disorder (TD) and autism (ASD), genes achieving TADA q<0.1 in ASD, genes with genome-wide significant statistical evidence for association with developmental disorders, and genes that are targets of CHD8 in the developing human brain. There was no significant enrichment for genes harboring de novo variants in intellectual disability (ID) or schizophrenia (SCZ), and no enrichment for any class of de novo variation in unaffected siblings in the SSC (Table 3, Table S6). Overlap between OCD and TD remained significant for all mutational classes, even when omitting variants from OCD subjects with comorbid tics (Table S6).
Table 3 –
Overlap between OCD de novo damaging mutations and gene sets
| LGD | Missense | Synonymous | ||||||||||
| Comparison gene seta | Obsb | Expc | O/Ed | Pe | Obs | Exp | O/E | P | Obs | Exp | O/E | P |
| TD | 5 | 0.29 | 17.4 | 2 × 10−5 | 14 | 1.33 | 10.6 | 1 × 10−5 | 3 | 0.81 | 3.72 | 0.047 |
| ASD | 9 | 4.55 | 1.98 | 0.037 | 25 | 17.94 | 1.39 | 0.045 | 9 | 8.08 | 1.11 | 0.42 |
| SCZ | 1 | 1.00 | 1.00 | 0.63 | 7 | 5.38 | 1.30 | 0.29 | 0 | 1.84 | 0 | 1 |
| DD | 2 | 2.78 | 0.72 | 0.77 | 5 | 6.50 | 0.77 | 0.79 | 4 | 2.27 | 1.76 | 0.19 |
| ID | 0 | 0.49 | 0 | 1 | 1 | 1.28 | 0.78 | 0.72 | 0 | 0.43 | 0 | 1 |
| Unaffected | 2 | 1.35 | 1.48 | 0.39 | 5 | 8.97 | 0.56 | 0.95 | 2 | 4.20 | 0.48 | 0.93 |
| Obs | Exp | O/E | P | |||||||||
| DD significantf | 4 | 0.84 | 4.76 | 0.010 | ||||||||
| ASD – q<0.1g | 3 | 0.65 | 4.64 | 0.027 | ||||||||
| CHD8 brainh | 20 | 13.03 | 1.54 | 0.030 | ||||||||
Comparisons for autism (ASD), schizophrenia (SCZ), developmental disorders (DD), Tourette’s disorder (TD), intellectual disability (ID), and unaffected siblings are between genes harboring de novo LGD, missense, or synonymous variants in these phenotypes and those harboring damaging (LGD or Mis-D) variants in OCD (n=89). Gene lists and their references are in Table S4.
Observed number of genes overlapping between sets.
Expected number of genes overlapping between sets, determined by 100,000 random mutation set simulations using DNENRICH (Fromer et al., 2014).
O/E, observed divided by expected number of genes overlapping between sets.
P-value is one-sided under a binomial model of greater than expected hits per gene set, calculated by DNENRICH using 100,000 permutations.
93 genes with genome-wide significant statistical evidence for association with developmental disorders (Deciphering Developmental Disorders Study, 2017).
65 ASD genes with False Discovery Rate (q) < 0.1 by TADA, considering data across exome sequencing studies (Sanders et al., 2015).
Exploratory pathway and network analyses
Using our list of genes harboring de novo damaging variants in OCD (n=89), we performed exploratory analyses to determine shared underlying canonical pathways and functional connectivity. Using the GeNets algorithm, OCD genes mapping onto a meta-network displayed significantly more connectivity than expected by chance (p=0.026) (Figure S6, Table S7). An additional 68 “tier 1” candidate genes were predicted by the GeNets algorithm, based on their high network connectivity to our original 89 input genes. These candidate genes are provided for reference in Table S7. All GeNets results for this analysis are also available in interactive form here: https://www.broadinstitute.org/genets#/visualize/58d9425ea4e00291af652379.
Based on the results from two pathway analysis tools, MetaCore and IPA, our input gene list is enriched for canonical pathways related to immune response, particularly the complement system (FDR 0.13). Other enriched canonical pathways include granulocyte-macrophage colony stimulating factor (GM-CSF) signaling, neurotrophin/tyrosine kinase signaling, B cell receptor signaling, and focal adhesion kinase signaling (Table S8). With regard to biological processes, sodium ion homeostasis shows the greatest enrichment using MetaCore (FDR 3.7 × 10−7). With regard to diseases, multiple cancer-related networks show the most enrichment (FDR ~10−8–10−9) using MetaCore and IPA.
DISCUSSION
By whole-exome sequencing of OCD parent-child trios, we have demonstrated a strong association between de novo damaging (LGD and Mis-D) coding variants and OCD cases (Table 1, Figure 2). As seen in studies of other neurodevelopmental disorders, these results can be leveraged to systematically identify OCD risk genes. In the current study, two genes, CHD8 and SCUBE1, have an FDR q<0.1, meeting criteria for high-confidence association with OCD (Table 2).
Of the subjects with predicted damaging de novo mutations in CHD8 (Table S2), subject OCD8015.p1 was diagnosed with OCD and hair-pulling disorder (trichotillomania); subject OCD8134.p1 was diagnosed with OCD, Tourette’s disorder, ADHD, and separation anxiety disorder. Of the subjects with predicted damaging de novo mutations in SCUBE1, subject 8100.p1 was diagnosed only with OCD, and subject OCD8141.p1 was diagnosed with OCD and Tourette’s disorder. Based on a structured clinical interview, no subjects in this study had a diagnosis of autism spectrum disorder or intellectual disability. The presence of Tourette’s disorder in one subject each with a CHD8 and SCUBE1 predicted damaging de novo mutation raises the question of whether these genes may play a role in the Tourette phenotype. Clinically, OCD and Tourette have high rates of comorbidity (45), and our genetic overlap analysis (Table 3, Table S6) supports the likelihood of shared genetic risk. On the other hand, the largest WES study of 802 Tourette’s disorder parent-child trios (including 37% with comorbid OCD) (24) did not find evidence for CHD8 and SCUBE1 as risk genes. Continued WES of trios recruited for OCD and for Tourette’s disorder, currently underway, is likely to clarify the relative contribution of these genes to each disorder. Future studies should also attempt more extensive phenotyping of these patients with predicted damaging mutations in CHD8 and SCUBE1.
SCUBE1 has not been extensively studied. While it is expressed in the developing brain and nervous system (46, 47), functional studies to date have focused mostly on its potential role in platelet activation and adhesion (48, 49). A study in mice has shown downregulated SCUBE1 expression in response to inflammatory stimuli (47), but this gene has not yet been implicated in disorders of the brain or nervous system. Interestingly, increased levels of pro-inflammatory markers have been reported in several studies of children and adolescents with OCD (50–53).
On the other hand, there are several recent and ongoing studies of CHD8, a gene that has emerged as having the strongest association with autism spectrum disorder via the identification of multiple de novo LGD variants in unrelated parent-child trios (Figure S5) (39, 54–56). CHD8 is highly expressed in the developing brain (57). It encodes an ATP-dependent chromatin remodeler that binds to tri-methylated histone H3 lysine 4, a post-translational histone modification present at active promoters (58–60). Loss of CHD8 function appears to contribute to autism pathology by disrupting the expression of its target genes, which are themselves enriched for high confidence autism risk genes (57). While OCD subjects with de novo damaging CHD8 variants in our study do not meet any diagnostic criteria for autism, this finding suggests there may be overlapping biological mechanisms between the two disorders and leads us to hypothesize that genes regulated by CHD8 may similarly be enriched for OCD risk genes. Indeed, we see significant overlap between our OCD genes and ASD genes, as well as CHD8 gene targets mapped in the developing human brain (57) (Table 3, Table S6).
While the majority of CHD8 case reports in the literature do not mention OCD traits, Talkowski et al. reported the case of a patient diagnosed with ASD, intellectual disability and OCD in the context of CHD8 disruption by a de novo balanced translocation (61). Three cases reported by Bernier et al. mention repetitive motor movements, rare repetitive behaviors, repetitive play, and repetitive/scripted speech; four cases mention problems with anxiety (55). Repetitive behaviors and increased anxiety have been reported in Chd8 haploinsufficient mice (62). One hypothesis from these observations is that CHD8 may either directly or indirectly alter sensorimotor gating which underlies multiple phenotypes, including OCD, ASD, anxiety, and tics (63). Further investigation of CHD8 in OCD and more extensive phenotyping of OCD patients with CHD8 mutations will be important to gain further insight into the mechanisms underlying association.
Based on data from this study, we estimate that 34% of de novo damaging mutations seen in OCD carry risk and that 335 genes confer risk in 22% of patients (Figure 2B). Given our OCD sample size, the 95% confidence intervals around these contribution estimates are wide and need refinement by continued sequencing of OCD trios.
Mindful of the fact that more than half of genes harboring de novo damaging variants in our study may not be true risk genes, we consider our pathway and network analyses as exploratory at this stage. Nevertheless, we see preliminary evidence that genes identified by de novo damaging variants in OCD are functionally connected to a greater degree than expected by chance (Figure S6, Table S7). Furthermore, these genes may be enriched in immunological and complement system canonical pathways (FDR 0.13; Table S8), consistent with our pilot study of exome sequencing in 20 OCD trios (31). More robust enrichment is seen for sodium ion homeostasis processes (FDR~10−7) and cancer-related disease pathways (FDR~10−8–10−9). These analyses should be viewed as preliminary and should be repeated as more high-confidence OCD risk genes are identified.
While not rising to the level of a high-confidence risk gene in this study, it is notable that we identified an OCD de novo damaging (Mis-D) variant in DLGAP1 (discs, large homolog-associated protein 1). In a genome-wide association study by the International OCD Foundation Genetic Collaborative (IOCDF-GC), the lowest p-values for their case-control analysis were found for two SNPs located within DLGAP1 (2.49×10−6, 3.44×10−6) (20). A subsequent GWAS by the OCD Collaborative Genetics Study (OCGAS) identified a SNP nearby this gene with a prominent signal (p=2.67×10−4) (18). Furthermore, a rare paternally-inherited duplication in DLGAP1 was recently reported in a child with OCD, Tourette syndrome, and anxiety (64). DLGAP1 is a member of the neuronal postsynaptic density complex and is in the same family as DLGAP3 (SAPAP3), a gene associated with OCD-like behaviors in a knockout mouse model (65). Therefore, evidence is beginning to converge on this gene as one of great interest in OCD genetics.
Successful gene discovery by leveraging gene-level recurrent de novo variation in autism, where over 65 genes have now been identified (26, 28, 29), and the results presented here for OCD, strongly reinforce the value of continuing WES in larger cohorts of OCD parent- child trios. Our models predict that by increasing the sample size of this study to 500 trios, we will gain 9 additional high-confidence risk genes and 22 probable risk genes. Further increasing to 1,000 trios will yield a total of 40 high-confidence and 77 probable risk genes. Discovering risk genes will change the status quo in OCD genetics by allowing new studies in model systems (e.g. animal models, induced pluripotent stem cells) and network analyses. Such studies will provide insights into OCD pathophysiology that are critical prerequisites for the discovery of novel therapeutic targets to alleviate the suffering of those with OCD.
Supplementary Material
Table S4 – Distribution of inherited coding variants in OCD cases and controls
Figure S1 – PCA scree and individual plots.
Scree plots following Principal Components Analysis (PCA), showing (A) the percentage of variance captured by each of the first 32 principal components, and (B) the cumulative percentage of variance captured by these same components in the exome metrics data from cases and controls. The “elbow” of the scree plot is visualized to be around the 5th principal component. This was confirmed by the Factominer R code function “estim_ncp()”. The first 5 PCs capture almost 80% of the variance, and this number of PCs was used to determine PCA outliers during quality control (see Table S1 and Supplementary Methods). (C) Individual plots for the first two principal components, based on PCA of exome sequencing quality metrics. OCD cases are plotted in red, and controls in blue. The first two PCs together capture 50.1% of the variance. R code to generate this data and figure are in Supplementary Methods, and individual PC factor values are in Table S1. This figure includes PCA outliers (>3 standard deviations from the mean in PCs 1–5), which were removed during quality control, prior to further analysis of case-control data.
Figure S2 – Maximum Likelihood Estimate (MLE) of number of OCD risk genes.
Assuming each number of possible risk genes between 1–2,500, 100,000 simulations were conducted to determine the number of risk genes that yielded the closest agreement between our observed and simulated data. In each simulation, we generated 95 variants (the number of de novo damaging variants observed in our OCD sample), then randomly assigned a percentage of variants (determined by the fraction of de novo damaging variants estimated to carry OCD risk) to the risk genes, recording the frequency at which the number of genes with two and three recurrent variants matched the number observed in our study (2 and 1, respectively). This MLE method yields an estimate of 335 OCD risk genes (red vertical line), a number that is in close agreement with that from an alternative "unseen species" method (317 genes, 95% CI: 190–454).
Figure S3 – Gene discovery by number of trios sequenced.
Using the MLE estimate of 335 risk genes, we estimated the number of probable (q<0.3) and high-confidence (q<0.1) risk genes that will be discovered as more OCD trios are sequenced. We performed 10,000 simulations at each cohort size from 25–3,000 trios, randomly generating variants and assigning to risk genes in agreement with the proportions seen in our data, then applying the TADA-Denovo algorithm.
Figure S4 – Sequencing coverage and parental age distributions.
Distribution boxplots of values for (A) mean target coverage, (B) median target coverage, (C) number of base pairs in all “callable” regions, (D) number of base pairs in coding “callable” regions, (E) paternal age, and (F) maternal age for both OCD and control cohorts. For each cohort, the box extends from the first through third quartiles, and the horizontal line is at the second quartile (median) of the data. Whiskers extend to the largest non-outliers, and outlier data points are plotted individually. For each comparison, a p-value was calculated using a two-sided Wilcoxon rank sum test with continuity correction. Panels A-D show increased opportunity for variant calling in the OCD cohort, necessitating the use of de novo mutation rate comparisons within the callable exome, as explained in the main text and methods. Panels E-F show no significant difference in parental ages between case and control cohorts. Also see Table S1.
Figure S5 – CHD8 variants in OCD and ASD.
Two de novo likely gene disrupting (LGD, red) and damaging missense (Mis-D, black) variants identified in CHD8 among OCD probands are indicated. ASD-associated de novo LGD and Mis-D mutations reported in the Simons Foundation Autism Research Initiative (SFARI) database (accessed April 12, 2017) are also shown in muted colors. Only variants with identifiable allele or residue sequence positions in the SFARI database were included in the above protein diagram, and splice site variants across cohorts are indicated by the respective allele change and underlined. Annotated protein domains predicted with confidence by the Simple Modular Architecture Research Tool (SMART) are shown as follows: CHR, chromatin organization modifier domain (blue), DEX, DEAD-like helicases superfamily (yellow), HELC, helicase superfamily c-terminal domain (pink), and BRK, domain of unknown function associated with CHROMO domain helicases (purple).
Figure S6 – GeNets network analysis without candidates.
Using the GeNets algorithm (https://apps.broadinstitute.org/genets), we mapped all 89 genes harboring de novo damaging mutations in OCD (excluding two genes, TTN and CACNA1E, which harbored de novo damaging variants in control subjects) onto the GeNets Metanetwork v1.0 to determine whether they are functionally connected. The density of the mapped network (density = number of edges / number of possible edges) was greater than 95% of randomly sampled gene sets, indicating that the network is significantly more connected than random (p=0.026). In the figure, node (gene) size is proportional to the number of connections. Node color indicates “community” assignment. A community is a set of genes that are more connected to one another than to another group of genes. Interactive results are available here: https://www.broadinstitute.org/genets#/visualize/58d9425ea4e00291af652379.
Table S1 – Phenotype, exome sequencing metrics, and principal components analysis.
First tab contains individual-level sample information (columns A-K), including family ID, individual ID, phenotype, cohort, collection site, gender, capture platform, size of “callable exome”, and parental age (years) at birth, where available. Column L lists reasons for any sample exclusions by quality control methods; “0” indicates that the sample was not excluded and was included in subsequent analyses. Columns M-AH list individual sample sequencing metrics generated using PicardTools, and GATK DepthOfCoverage tools. Columns AI-AS list individual sample sequencing metrics generated using PLINK/SEQ (i-stats; https://psychgen.u.hpc.mssm.edu/plinkseq/stats.shtml). Columns B, M-AS were included in Principal Components Analysis (PCA). Third tab contains cohort-level metrics calculated using samples passing quality control. ±95% confidence intervals are given, when applicable. Fourth tab contains coordinates generated for each sample for the top 10 principal components following PCA. The code used to generate this data is included in Supplementary Methods. Using these coordinates, we removed trios with family members falling more than three standard deviations from the mean in any of the first five principal components; this information is contained in the fifth tab.
Table S2 – Annotated de novo variants in OCD and controls.
Detailed information on all high confidence de novo variants in cases and controls. These variants were annotated using Annovar, based on RefSeq hg19 gene definitions. Column descriptions are provided in a separate tab of this file. A third tab provides the number of each de novo variant type per sample.
Table S3 – Annotated inherited variants in OCD and controls.
Detailed information on all high confidence inherited variants in cases and controls. These variants were annotated using Annovar, based on RefSeq hg19 gene definitions. Column descriptions are provided in a separate tab of this file. A third tab provides the number of each inherited variant type per sample.
Table S5 – Gene-level de novo mutation rates, variant counts, and TADA results.
First tab contains de novo mutation rates used to perform subsequent maximum likelihood estimation (MLE) and TADA analyses. The following mutation rates are listed for each gene: overall, likely gene disrupting (lgd), predicted damaging missense (misD), and all damaging (lgd + misD). These mutation rates were previously published (Ware et al., 2015) from unaffected parent-child trios. The code used to generate the mutation rate table is provided in Supplementary Methods. Second tab contains the input file for the TADA-Denovo code. Gene-level expected mutation rates for LGD (“mut.cls1” column) and Mis-D variants (“mut.cls2” column) are listed, along with their respective observed de novo mutation counts in our OCD data (“dn.cls1” and “dn.cls2”, respectively). Code for running TADA-Denovo is given in Supplementary Methods. Third tab contains the final output results from TADA-Denovo code provided in Supplementary Methods. Genes harboring more than one damaging de novo (LGD or Mis-D) variant in OCD probands are highlighted in yellow (SCUBE1, CHD8, TTN). Two of these genes (SCUBE1 and CHD8) exceeded thresholds for being considered a probable (qval < 0.3) or high confidence (qval < 0.1) risk gene. Fourth tab contains the input file for the TADA (Denovo + Inherited) code. Gene-level expected mutation rates for LGD (“mut.cls1” column) and Mis-D variants (“mut.cls2” column) are listed, along with their respective observed de novo mutation counts in our OCD data (“dn.cls1” and “dn.cls2”, respectively), observed transmitted mutation counts (trans.cls1, trans.cls2), and observed non-transmitted mutation counts (present in either parent but not the child; ntrans.cls1, ntrans.cls2). Code for running TADA (Denovo + Inherited) is given in Supplementary Methods.
Table S6 – DNENRICH gene lists and results.
See Supplementary Methods for details of DNENRICH analysis. First tab contains the input gene lists to determine enrichment for our OCD damaging de novo mutations. Second tab contains input for DNENRICH analysis. Each row represents a de novo damaging mutation in an OCD proband. Third tab contains final results output from the DNENRICH primary analysis, also shown in Table 3. Fourth tab contains input for DNENRICH secondary analysis; each row represents a de novo damaging mutation in an OCD proband without known tics or Tourette’s disorder. Fifth tab contains the results of this secondary analysis. Significantly enriched gene sets are highlighted.
Table S7 – GeNets network connectivity analysis results.
Complete results from GeNets network analysis of de novo damaging variants found in OCD probands. First tab contains summary statistics of the resulting network, considered both with and without nearby predicted “tier 1” candidate genes. Second tab contains the input gene list and the candidate genes predicted by the network analysis. Third tab groups genes (without predicted candidates) into nearby “communities” that are more connected with each other than their neighbors. Fourth tab contains network edges without the predicted candidates. Fifth tab contains gene community groupings, including predicted candidates. Sixth tab contains network edges including predicted candidates. See Methods for further details of this analysis.
Table S8 – MetaCore and Ingenuity Pathway Analysis (IPA) gene enrichment analysis results.
Complete results from Metacore (first tab) and IPA (second tab) gene enrichment analyses, with p-values calculated by each analysis algorithm. See Methods for details of these analyses.
ACKNOWLEDGEMENTS
We wish to thank the families who have participated in and contributed to this study. Control subject data were obtained from the NIH-supported National Database for Autism Research (NDAR). NDAR is a collaborative informatics system created by the National Institutes of Health to provide a national resource to support and accelerate research in autism. Dataset identifier: 2042. This manuscript content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or of the Submitters submitting original data to NDAR. Additionally, we wish to thank the Tourette International Collaborative Genetics Study for contributing published genetic data. Research reported in this application was supported by grants from the Allison Family Foundation and the National Institute of Mental Health of the National Institutes of Health under award number R01MH114927 [TVF]; FAPESP (process number: 2014/01585–5) and CNPq (process number: 460928/2014–7) [CC]; the Ontario Mental Health Foundation (OMHF) and private donations from the Frederick W. Thompson family [MAR, JLK]. An earlier version of the manuscript was published on the bioRxiv preprint sever: https://doi.org/10.1101/127712.
Footnotes
DISCLOSURES
JLK is a Scientific Advisory Board member of AssureRx. JLK has received speaker honoraria and expenses from Eli Lilly and Novartis, consultant honoraria and expenses from Roche, and expenses from AssureRx. MAR has received research support through grants from Roche and speaker honoraria from Lundbeck. TVF has received research support from Shire, the Simons Foundation, and the National Institute of Mental Health. All other authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Fontenelle LF, Mendlowicz MV, Versiani M (2006): The descriptive epidemiology of obsessive-compulsive disorder. Progress in neuro-psychopharmacology & biological psychiatry. 30:327–337. [DOI] [PubMed] [Google Scholar]
- 2.Karno M, Golding JM, Sorenson SB, Burnam MA (1988): The epidemiology of obsessive-compulsive disorder in five US communities. Arch Gen Psychiatry. 45:1094–1099. [DOI] [PubMed] [Google Scholar]
- 3.Ruscio AM, Stein DJ, Chiu WT, Kessler RC (2010): The epidemiology of obsessive-compulsive disorder in the National Comorbidity Survey Replication. Mol Psychiatry. 15:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weissman MM, Bland RC, Canino GJ, Greenwald S, Hwu HG, Lee CK, et al. (1994): The cross national epidemiology of obsessive compulsive disorder. The Cross National Collaborative Group. J Clin Psychiatry. 55 Suppl:5–10. [PubMed] [Google Scholar]
- 5.Torres AR, Prince MJ, Bebbington PE, Bhugra D, Brugha TS, Farrell M, et al. (2006): Obsessive-compulsive disorder: prevalence, comorbidity, impact, and help-seeking in the British National Psychiatric Morbidity Survey of 2000. Am J Psychiatry. 163:1978–1985. [DOI] [PubMed] [Google Scholar]
- 6.Miguel EC, do Rosario-Campos MC, Prado HS, do Valle R, Rauch SL, Coffey BJ, et al. (2000): Sensory phenomena in obsessive-compulsive disorder and Tourette’s disorder. J Clin Psychiatry. 61:150–156; quiz 157. [DOI] [PubMed] [Google Scholar]
- 7.Shavitt RG, de Mathis MA, Oki F, Ferrao YA, Fontenelle LF, Torres AR, et al. (2014): Phenomenology of OCD: lessons from a large multicenter study and implications for ICD-11. J Psychiatr Res. 57:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veale D, Roberts A (2014): Obsessive-compulsive disorder. BMJ (Clinical research ed). 348:g2183. [DOI] [PubMed] [Google Scholar]
- 9.Bobes J, Gonzalez MP, Bascaran MT, Arango C, Saiz PA, Bousono M (2001): Quality of life and disability in patients with obsessive-compulsive disorder. Eur Psychiatry. 16:239–245. [DOI] [PubMed] [Google Scholar]
- 10.Meier SM, Mattheisen M, Mors O, Schendel DE, Mortensen PB, Plessen KJ (2016): Mortality Among Persons With Obsessive-Compulsive Disorder in Denmark. JAMA psychiatry. 73:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lochner C, Fineberg NA, Zohar J, van Ameringen M, Juven-Wetzler A, Altamura AC, et al. (2014): Comorbidity in obsessive-compulsive disorder (OCD): a report from the International College of Obsessive-Compulsive Spectrum Disorders (ICOCS). Comprehensive psychiatry. 55:1513–1519. [DOI] [PubMed] [Google Scholar]
- 12.Skoog G, Skoog I (1999): A 40-year follow-up of patients with obsessive-compulsive disorder [see commetns]. Arch Gen Psychiatry. 56:121–127. [DOI] [PubMed] [Google Scholar]
- 13.Eisen JL, Sibrava NJ, Boisseau CL, Mancebo MC, Stout RL, Pinto A, et al. (2013): Five-year course of obsessive-compulsive disorder: predictors of remission and relapse. J Clin Psychiatry. 74:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monzani B, Rijsdijk F, Harris J, Mataix-Cols D (2014): The structure of genetic and environmental risk factors for dimensional representations of DSM-5 obsessive-compulsive spectrum disorders. JAMA psychiatry. 71:182–189. [DOI] [PubMed] [Google Scholar]
- 15.Pauls DL, Alsobrook JP, Goodman W, Rasmussen S, Leckman JF (1995): A family study of obsessive-compulsive disorder. Am J Psychiatry. 152:76–84. [DOI] [PubMed] [Google Scholar]
- 16.Taylor S (2011): Etiology of obsessions and compulsions: a meta-analysis and narrative review of twin studies. Clinical psychology review. 31:1361–1372. [DOI] [PubMed] [Google Scholar]
- 17.van Grootheest DS, Cath DC, Beekman AT, Boomsma DI (2005): Twin studies on obsessive-compulsive disorder: a review. Twin research and human genetics : the official journal of the International Society for Twin Studies. 8:450–458. [DOI] [PubMed] [Google Scholar]
- 18.Mattheisen M, Samuels JF, Wang Y, Greenberg BD, Fyer AJ, McCracken JT, et al. (2014): Genome-wide association study in obsessive-compulsive disorder: results from the OCGAS. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costas J, Carrera N, Alonso P, Gurriaran X, Segalas C, Real E, et al. (2016): Exon-focused genome-wide association study of obsessive-compulsive disorder and shared polygenic risk with schizophrenia. Transl Psychiatry. 6:e768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart SE, Yu D, Scharf JM, Neale BM, Fagerness JA, Mathews CA, et al. (2013): Genome-wide association study of obsessive-compulsive disorder. Mol Psychiatry. 18:788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deciphering Developmental Disorders Study (2017): Prevalence and architecture of de novo mutations in developmental disorders. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler EE, et al. (2013): De novo mutations in epileptic encephalopathies. Nature. 501:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, et al. (2012): Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 380:1674–1682. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Mandell JD, Kumar Y, Sun N, Morris MT, Arbelaez J, et al. (2018): De Novo Sequence and Copy Number Variants Are Strongly Associated with Tourette Disorder and Implicate Cell Polarity in Pathogenesis. Cell reports. 24:3441–3454.e3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willsey AJ, Fernandez TV, Yu D, King RA, Dietrich A, Xing J, et al. (2017): De Novo Coding Variants Are Strongly Associated with Tourette Disorder. Neuron. 94:486–499 e489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. (2014): Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 515:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong S, Walker MF, Carriero NJ, DiCola M, Willsey AJ, Ye AY, et al. (2014): De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell reports. 9:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. (2014): The contribution of de novo coding mutations to autism spectrum disorder. Nature. 515:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, et al. (2015): Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 87:1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, et al. (2013): Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 155:997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cappi C, Brentani H, Lima L, Sanders SJ, Zai G, Diniz BJ, et al. (2016): Whole-exome sequencing in obsessive-compulsive disorder identifies rare mutations in immunological and neurodevelopmental pathways. Transl Psychiatry. 6:e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.American Psychiatric Association (2000): Diagnostic and statistical manual of mental disorders : DSM-IV-Tr. Washington, DC: American Psychiatric Association. [Google Scholar]
- 33.American Psychiatric Association; (2013): Diagnostic and statistical manual of mental disorders : DSM-5. [Google Scholar]
- 34.Dietrich A, Fernandez TV, King RA, State MW, Tischfield JA, Hoekstra PJ, et al. (2015): The Tourette International Collaborative Genetics (TIC Genetics) study, finding the genes causing Tourette syndrome: objectives and methods. European Child & Adolescent Psychiatry. 24:141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.First MB, Spitzer RL, Gibbon M, Williams JB (2002): Structured clinical interview for DSM-IV-TR axis I disorders, research version, patient edition. SCID-I/P New York, NY. [Google Scholar]
- 36.Fischbach GD, Lord C (2010): The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 68:192–195. [DOI] [PubMed] [Google Scholar]
- 37.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. (2010): The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DnA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. (2016): Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. (2012): De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 485:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, et al. (2013): Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 9:e1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, et al. (2015): De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 350:1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. (2014): De novo mutations in schizophrenia implicate synaptic networks. Nature. 506:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deciphering Developmental Disorders Study (2015): Large-scale discovery of novel genetic causes of developmental disorders. Nature. 519:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeo G, Burge CB (2004): Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. Journal of computational biology : a journal of computational molecular cell biology. 11:377–394. [DOI] [PubMed] [Google Scholar]
- 45.Franklin ME, Harrison JP, Benavides KL (2012): Obsessive-compulsive and tic-related disorders. Child Adolesc Psychiatr Clin N Am. 21:555–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimmond S, Larder R, Van Hateren N, Siggers P, Hulsebos TJ, Arkell R, et al. (2000): Cloning, mapping, and expression analysis of a gene encoding a novel mammalian EGF-related protein (SCuBe1). Genomics. 70:74–81. [DOI] [PubMed] [Google Scholar]
- 47.Yang RB, Ng CK, Wasserman SM, Colman SD, Shenoy S, Mehraban F, et al. (2002): Identification of a novel family of cell-surface proteins expressed in human vascular endothelium. J Biol Chem. 277:46364–46373. [DOI] [PubMed] [Google Scholar]
- 48.Tu CF, Su YH, Huang YN, Tsai MT, Li LT, Chen YL, et al. (2006): Localization and characterization of a novel secreted protein SCUBE1 in human platelets. Cardiovascular research. 71:486–495. [DOI] [PubMed] [Google Scholar]
- 49.Wu MY, Lin YC, Liao WJ, Tu CF, Chen MH, Roffler SR, et al. (2014): Inhibition of the plasma SCUBE1, a novel platelet adhesive protein, protects mice against thrombosis. Arteriosclerosis, thrombosis, and vascular biology. 34:1390–1398. [DOI] [PubMed] [Google Scholar]
- 50.Gabbay V, Coffey BJ, Guttman LE, Gottlieb L, Katz Y, Babb JS, et al. (2009): A cytokine study in children and adolescents with Tourette’s disorder. Progress in neuro-psychopharmacology & biological psychiatry. 33:967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mittleman BB, Castellanos FX, Jacobsen LK, Rapoport JL, Swedo SE, Shearer GM (1997): Cerebrospinal fluid cytokines in pediatric neuropsychiatric disease. Journal of immunology (Baltimore, Md : 1950). 159:2994–2999. [PubMed] [Google Scholar]
- 52.Leckman JF, Katsovich L, Kawikova I, Lin H, Zhang H, Kronig H, et al. (2005): Increased serum levels of interleukin-12 and tumor necrosis factor-alpha in Tourette’s syndrome. Biol Psychiatry. 57:667–673. [DOI] [PubMed] [Google Scholar]
- 53.Mitchell RH, Goldstein BI (2014): Inflammation in children and adolescents with neuropsychiatric disorders: a systematic review. J Am Acad Child Adolesc Psychiatry. 53:274–296. [DOI] [PubMed] [Google Scholar]
- 54.Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, et al. (2012): Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 485:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, et al. (2014): Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 158:263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. (2012): Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 338:1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, et al. (2015): The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nature communications. 6:6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson BA, Tremblay V, Lin G, Bochar DA (2008): CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol Cell Biol. 28:3894–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan CC, Zhao X, Florens L, Swanson SK, Washburn MP, Hernandez N (2007): CHD8 associates with human Staf and contributes to efficient U6 RNA polymerase III transcription. Mol Cell Biol. 27:8729–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. (2007): High-resolution profiling of histone methylations in the human genome. Cell. 129:823–837. [DOI] [PubMed] [Google Scholar]
- 61.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, et al. (2012): Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 149:525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, et al. (2016): CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 537:675–679. [DOI] [PubMed] [Google Scholar]
- 63.Ahmari SE, Risbrough VB, Geyer MA, Simpson HB (2012): Impaired sensorimotor gating in unmedicated adults with obsessive-compulsive disorder. Neuropsychopharmacology. 37:1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gazzellone MJ, Zarrei M, Burton CL, Walker S, Uddin M, Shaheen SM, et al. (2016): Uncovering obsessive-compulsive disorder risk genes in a pediatric cohort by high-resolution analysis of copy number variation. Journal of neurodevelopmental disorders. 8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Welch JM, Lu J, Rodriguiz RM, Trotta NC, Peca J, Ding JD, et al. (2007): Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature. 448:894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4 – Distribution of inherited coding variants in OCD cases and controls
Figure S1 – PCA scree and individual plots.
Scree plots following Principal Components Analysis (PCA), showing (A) the percentage of variance captured by each of the first 32 principal components, and (B) the cumulative percentage of variance captured by these same components in the exome metrics data from cases and controls. The “elbow” of the scree plot is visualized to be around the 5th principal component. This was confirmed by the Factominer R code function “estim_ncp()”. The first 5 PCs capture almost 80% of the variance, and this number of PCs was used to determine PCA outliers during quality control (see Table S1 and Supplementary Methods). (C) Individual plots for the first two principal components, based on PCA of exome sequencing quality metrics. OCD cases are plotted in red, and controls in blue. The first two PCs together capture 50.1% of the variance. R code to generate this data and figure are in Supplementary Methods, and individual PC factor values are in Table S1. This figure includes PCA outliers (>3 standard deviations from the mean in PCs 1–5), which were removed during quality control, prior to further analysis of case-control data.
Figure S2 – Maximum Likelihood Estimate (MLE) of number of OCD risk genes.
Assuming each number of possible risk genes between 1–2,500, 100,000 simulations were conducted to determine the number of risk genes that yielded the closest agreement between our observed and simulated data. In each simulation, we generated 95 variants (the number of de novo damaging variants observed in our OCD sample), then randomly assigned a percentage of variants (determined by the fraction of de novo damaging variants estimated to carry OCD risk) to the risk genes, recording the frequency at which the number of genes with two and three recurrent variants matched the number observed in our study (2 and 1, respectively). This MLE method yields an estimate of 335 OCD risk genes (red vertical line), a number that is in close agreement with that from an alternative "unseen species" method (317 genes, 95% CI: 190–454).
Figure S3 – Gene discovery by number of trios sequenced.
Using the MLE estimate of 335 risk genes, we estimated the number of probable (q<0.3) and high-confidence (q<0.1) risk genes that will be discovered as more OCD trios are sequenced. We performed 10,000 simulations at each cohort size from 25–3,000 trios, randomly generating variants and assigning to risk genes in agreement with the proportions seen in our data, then applying the TADA-Denovo algorithm.
Figure S4 – Sequencing coverage and parental age distributions.
Distribution boxplots of values for (A) mean target coverage, (B) median target coverage, (C) number of base pairs in all “callable” regions, (D) number of base pairs in coding “callable” regions, (E) paternal age, and (F) maternal age for both OCD and control cohorts. For each cohort, the box extends from the first through third quartiles, and the horizontal line is at the second quartile (median) of the data. Whiskers extend to the largest non-outliers, and outlier data points are plotted individually. For each comparison, a p-value was calculated using a two-sided Wilcoxon rank sum test with continuity correction. Panels A-D show increased opportunity for variant calling in the OCD cohort, necessitating the use of de novo mutation rate comparisons within the callable exome, as explained in the main text and methods. Panels E-F show no significant difference in parental ages between case and control cohorts. Also see Table S1.
Figure S5 – CHD8 variants in OCD and ASD.
Two de novo likely gene disrupting (LGD, red) and damaging missense (Mis-D, black) variants identified in CHD8 among OCD probands are indicated. ASD-associated de novo LGD and Mis-D mutations reported in the Simons Foundation Autism Research Initiative (SFARI) database (accessed April 12, 2017) are also shown in muted colors. Only variants with identifiable allele or residue sequence positions in the SFARI database were included in the above protein diagram, and splice site variants across cohorts are indicated by the respective allele change and underlined. Annotated protein domains predicted with confidence by the Simple Modular Architecture Research Tool (SMART) are shown as follows: CHR, chromatin organization modifier domain (blue), DEX, DEAD-like helicases superfamily (yellow), HELC, helicase superfamily c-terminal domain (pink), and BRK, domain of unknown function associated with CHROMO domain helicases (purple).
Figure S6 – GeNets network analysis without candidates.
Using the GeNets algorithm (https://apps.broadinstitute.org/genets), we mapped all 89 genes harboring de novo damaging mutations in OCD (excluding two genes, TTN and CACNA1E, which harbored de novo damaging variants in control subjects) onto the GeNets Metanetwork v1.0 to determine whether they are functionally connected. The density of the mapped network (density = number of edges / number of possible edges) was greater than 95% of randomly sampled gene sets, indicating that the network is significantly more connected than random (p=0.026). In the figure, node (gene) size is proportional to the number of connections. Node color indicates “community” assignment. A community is a set of genes that are more connected to one another than to another group of genes. Interactive results are available here: https://www.broadinstitute.org/genets#/visualize/58d9425ea4e00291af652379.
Table S1 – Phenotype, exome sequencing metrics, and principal components analysis.
First tab contains individual-level sample information (columns A-K), including family ID, individual ID, phenotype, cohort, collection site, gender, capture platform, size of “callable exome”, and parental age (years) at birth, where available. Column L lists reasons for any sample exclusions by quality control methods; “0” indicates that the sample was not excluded and was included in subsequent analyses. Columns M-AH list individual sample sequencing metrics generated using PicardTools, and GATK DepthOfCoverage tools. Columns AI-AS list individual sample sequencing metrics generated using PLINK/SEQ (i-stats; https://psychgen.u.hpc.mssm.edu/plinkseq/stats.shtml). Columns B, M-AS were included in Principal Components Analysis (PCA). Third tab contains cohort-level metrics calculated using samples passing quality control. ±95% confidence intervals are given, when applicable. Fourth tab contains coordinates generated for each sample for the top 10 principal components following PCA. The code used to generate this data is included in Supplementary Methods. Using these coordinates, we removed trios with family members falling more than three standard deviations from the mean in any of the first five principal components; this information is contained in the fifth tab.
Table S2 – Annotated de novo variants in OCD and controls.
Detailed information on all high confidence de novo variants in cases and controls. These variants were annotated using Annovar, based on RefSeq hg19 gene definitions. Column descriptions are provided in a separate tab of this file. A third tab provides the number of each de novo variant type per sample.
Table S3 – Annotated inherited variants in OCD and controls.
Detailed information on all high confidence inherited variants in cases and controls. These variants were annotated using Annovar, based on RefSeq hg19 gene definitions. Column descriptions are provided in a separate tab of this file. A third tab provides the number of each inherited variant type per sample.
Table S5 – Gene-level de novo mutation rates, variant counts, and TADA results.
First tab contains de novo mutation rates used to perform subsequent maximum likelihood estimation (MLE) and TADA analyses. The following mutation rates are listed for each gene: overall, likely gene disrupting (lgd), predicted damaging missense (misD), and all damaging (lgd + misD). These mutation rates were previously published (Ware et al., 2015) from unaffected parent-child trios. The code used to generate the mutation rate table is provided in Supplementary Methods. Second tab contains the input file for the TADA-Denovo code. Gene-level expected mutation rates for LGD (“mut.cls1” column) and Mis-D variants (“mut.cls2” column) are listed, along with their respective observed de novo mutation counts in our OCD data (“dn.cls1” and “dn.cls2”, respectively). Code for running TADA-Denovo is given in Supplementary Methods. Third tab contains the final output results from TADA-Denovo code provided in Supplementary Methods. Genes harboring more than one damaging de novo (LGD or Mis-D) variant in OCD probands are highlighted in yellow (SCUBE1, CHD8, TTN). Two of these genes (SCUBE1 and CHD8) exceeded thresholds for being considered a probable (qval < 0.3) or high confidence (qval < 0.1) risk gene. Fourth tab contains the input file for the TADA (Denovo + Inherited) code. Gene-level expected mutation rates for LGD (“mut.cls1” column) and Mis-D variants (“mut.cls2” column) are listed, along with their respective observed de novo mutation counts in our OCD data (“dn.cls1” and “dn.cls2”, respectively), observed transmitted mutation counts (trans.cls1, trans.cls2), and observed non-transmitted mutation counts (present in either parent but not the child; ntrans.cls1, ntrans.cls2). Code for running TADA (Denovo + Inherited) is given in Supplementary Methods.
Table S6 – DNENRICH gene lists and results.
See Supplementary Methods for details of DNENRICH analysis. First tab contains the input gene lists to determine enrichment for our OCD damaging de novo mutations. Second tab contains input for DNENRICH analysis. Each row represents a de novo damaging mutation in an OCD proband. Third tab contains final results output from the DNENRICH primary analysis, also shown in Table 3. Fourth tab contains input for DNENRICH secondary analysis; each row represents a de novo damaging mutation in an OCD proband without known tics or Tourette’s disorder. Fifth tab contains the results of this secondary analysis. Significantly enriched gene sets are highlighted.
Table S7 – GeNets network connectivity analysis results.
Complete results from GeNets network analysis of de novo damaging variants found in OCD probands. First tab contains summary statistics of the resulting network, considered both with and without nearby predicted “tier 1” candidate genes. Second tab contains the input gene list and the candidate genes predicted by the network analysis. Third tab groups genes (without predicted candidates) into nearby “communities” that are more connected with each other than their neighbors. Fourth tab contains network edges without the predicted candidates. Fifth tab contains gene community groupings, including predicted candidates. Sixth tab contains network edges including predicted candidates. See Methods for further details of this analysis.
Table S8 – MetaCore and Ingenuity Pathway Analysis (IPA) gene enrichment analysis results.
Complete results from Metacore (first tab) and IPA (second tab) gene enrichment analyses, with p-values calculated by each analysis algorithm. See Methods for details of these analyses.