Figure 1.
Generation and Characterization of iMg
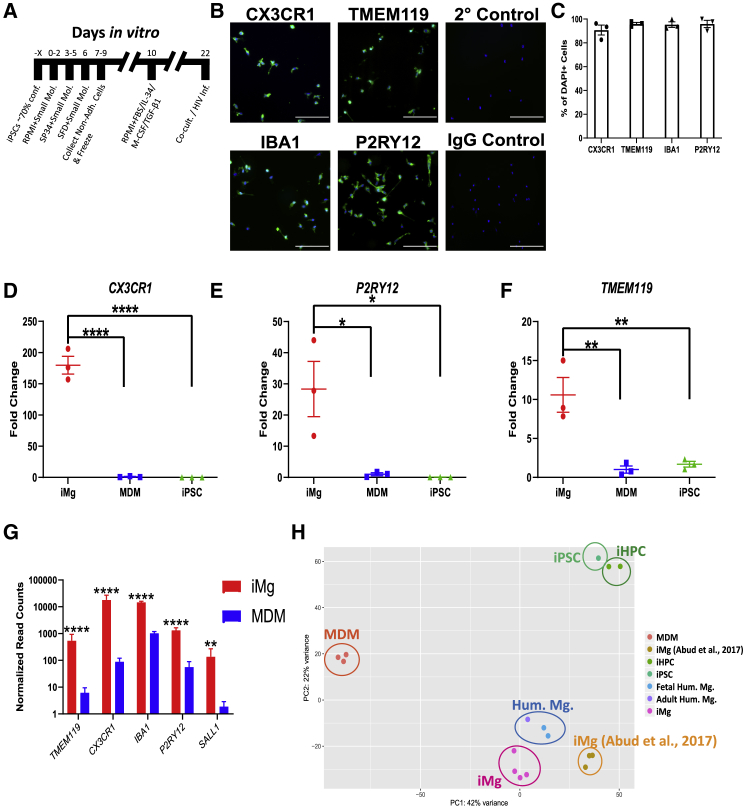
(A) Timeline for iMg differentiation.
(B) Immunostaining for microglia specific markers: CX3CR1, TMEM119, IBA1, and P2RY12, secondary and IgG controls. All cultures were stained for DAPI. Scale bar represents 50 μm.
(C) Percentage of DAPI(+) cells that are double-positive for microglia marker. n = 3 cell lines, error bars represent SEM.
(D–F) qRT-PCR validation for expression of CX3CR1 (D), P2RY12 (E), and TMEM119 (F) in iMg. Probes normalized to GAPDH expression. n = 3 cell lines, one-way ANOVA, Dunnett's post hoc analysis, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001; error bars represent SEM.
(G) Bulk RNA-seq normalized read counts of specific microglia markers from iMg and MDMs. n = 4 iMg lines, n = 3 MDMs. Benjamini-Hochberg, false discovery rate (FDR) = 0.01, ∗∗p < 0.01, ∗∗∗∗p < 0.0001; error bars represent SEM.
(H) Principal-component analysis (PCA) for monocyte-derived macrophages, iMicroglia, and (Abud et al., 2017) iMg, iPSC, iHPC, and primary human microglia. Each dot represents a bulk RNA-seq sample. MDM, monocyte-derived macrophage; iHPC, induced hematopoietic progenitor cells. iPSC, iHPC, Hum. Mg., and iMg samples were all retrieved from Abud et al. (2017).