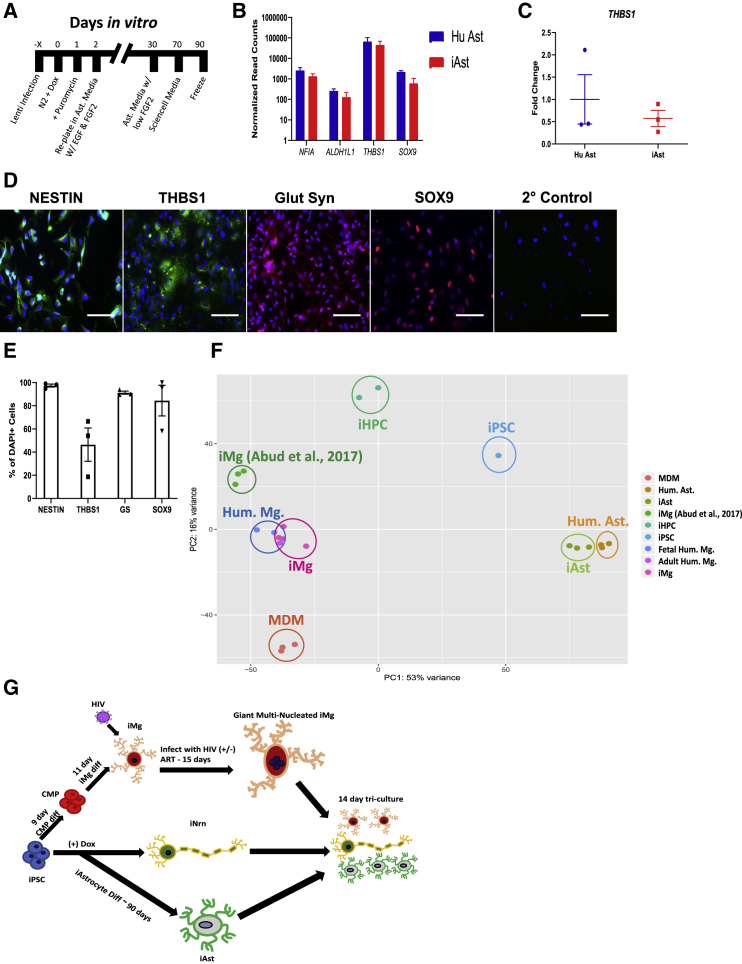
Figure 3.
Generation and Characterization of iAst and Formation of Tri-cultures
(A) Timeline for iAstrocyte differentiation.
(B) Bulk RNA-seq normalized read counts of select astrocyte specific genes from primary human astrocytes and iAst. n = 3 cell lines, Benjamini-Hochberg, FDR = 0.01, error bars represent SEM.
(C) qRT-PCR validation for expression of THBS1 in Hu Ast and iAst. Probe normalized to GAPDH expression. n = 3 cell lines, two-tailed t test, error bars represent SEM.
(D) Immunostaining for the astrocyte-specific markers: Nestin, thrombospondin-1, glutamine synthetase, SOX9, and secondary control. Scale bar represents 50 μm.
(E) Percentage of DAPI(+) cells that are double-positive for astrocyte marker. n = 3 cell lines, error bars represent SEM.
(F) PCA for monocyte-derived macrophages, iMicroglia, primary human astrocytes, and iAstrocytes, and (Abud et al., 2017) iMg, iPSC, iHPC, and primary human microglia. Each dot represents a bulk RNA-seq sample. MDM, monocyte-derived macrophage; Hum. Ast., primary fetal human astrocytes; iHPC, induced hematopoietic progenitor cells. iPSC, iHPC, Hum. Mg., iMg samples were all retrieved from Abud et al. (2017).
(G) Flowchart for differentiations into iMg, iAst, and iNrn and combination into tri-culture ± HIV infection and ART.