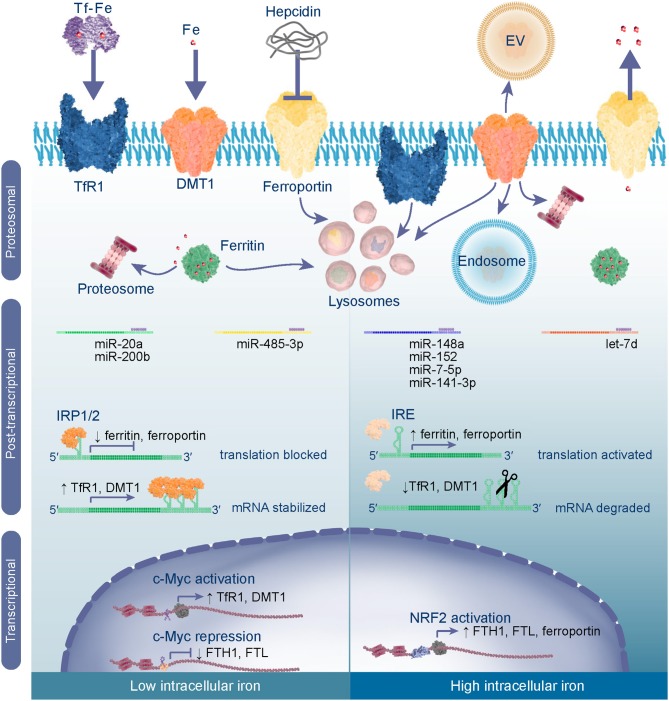
Figure 2.
Iron metabolism is regulated through transcriptional, post-transcriptional and proteosomal mechanisms. In low intracellular iron conditions c-Myc acts as a transcriptional activator of iron import genes TfR1 and DMT1 and represses expression of ferritin to increase the intracellular labile iron pool (LIP). In the cytoplasm iron regulatory proteins 1 and 2 (IRP1/IRP2) bind to iron responsive elements (IREs) in the 5′-UTR of ferritin and ferroportin mRNAs blocking their translation, whilst binding to 3′-UTR IREs stabilizes TfR1 and DMT1 mRNAs ensuring their translation. mRNAs may also be subject to post-transcriptional control by specific microRNAs (miRs) which bind to the 3′-UTR to inhibit translation or induce degradation of the transcript. To reduce systemic iron levels hepcidin is released by liver cells and targets ferroportin for lysosomal degradation thereby reducing export of iron into the blood stream. Degradation of ferritin is a mechanism for controlling intracellular labile iron levels by undergoing proteosomal or lysosomal degradation, to liberate iron from the nanocage and reduce apo-protein levels. When intracellular iron is high and oxidative stress is imminent NRF2 activates transcription of ferritin and ferroportin genes. In this case the IRPs are degraded and, hence, their translation is activated, whilst TfR1 and DMT1 mRNAs undergo endonuclease attack or are downregulated by miRs. Excess iron is stored in ferritin or exported from the cell via ferroportin. Further iron import is inhibited by degradation of TfR1 and DMT1 proteins or release from the cell membrane [internalization in the endosome or release in extracellular vesicles (EV)].